Click here to download this notebook.
How to use a xarray Dataset as NadirData source
NadirData can be created using anything than can be opened as a xarray dataset and compute statistics on it in the exact same manner. It allows to dynamically create a dataset by using as many inputs as you want, align their coordinates if needed, arrange them however you want and use it has your data source [doc].
Example: JASON 3 - 20hz netCDF
[1]:
import xarray as xr
from casys import NadirData, CasysPlot, Field, PlotParams
NadirData.enable_loginfo()
Create your source xarray dataset
[2]:
# Open with xarray the product files of Jason-3 (Track 1 to 9 of cycle 127)
ds = xr.open_mfdataset(
"/data/OCTANT_NG/cvi/JA3/JA3_GPN_2PdP127_00*.nc", combine="by_coords"
)
[3]:
ds["alt_echo_type"]
[3]:
<xarray.DataArray 'alt_echo_type' (time: 30130)> Size: 121kB
dask.array<concatenate, shape=(30130,), dtype=float32, chunksize=(3432,), chunktype=numpy.ndarray>
Coordinates:
* time (time) datetime64[ns] 241kB 2019-07-20T19:23:06.823509248 ... 20...
lat (time) float64 241kB dask.array<chunksize=(3411,), meta=np.ndarray>
lon (time) float64 241kB dask.array<chunksize=(3411,), meta=np.ndarray>
Attributes:
long_name: altimeter echo type
flag_values: [0 1]
flag_meanings: ocean_like non_ocean_like
comment: The altimeter echo type is determined by testing the rms ...Create your NadirData
[4]:
# Create your NadirData object
selection = "is_bounded(-40, deg_normalize(-90, lat_20hz), 60) && is_bounded(-50, deg_normalize(-180, lon_20hz), 150)"
cd_ncdf = NadirData(
source=ds,
select_clip=selection,
time="time_20hz",
longitude="lon_20hz",
latitude="lat_20hz",
)
[5]:
cd_ncdf.show_fields(containing="swh_20hz")
[5]:
| Name | Description | Unit |
|---|---|---|
| swh_20hz_ku | 20 Hz Ku band corrected significant waveheight All instrumental corrections included, i.e. modeled instrumental errors correction (modeled_instr_corr_swh_ku) and system bias | m |
| swh_20hz_c | 20 Hz C band corrected significant waveheight All instrumental corrections included, i.e. modeled instrumental errors correction (modeled_instr_corr_swh_c) and system bias | m |
| swh_20hz_ku_mle3 | 20 Hz Ku band corrected significant waveheight (MLE3 retracking) All instrumental corrections included, i.e. modeled instrumental errors correction (modeled_instr_corr_swh_ku_mle3) and system bias | m |
Play with it !
Add fields created with clips:
[6]:
f1 = Field(name="f1", source="swh_20hz_ku * swh_20hz_c + 3")
cd_ncdf.add_raw_data(name="F1", field=f1)
cd_ncdf.compute()
2025-05-14 10:59:48 INFO Reading ['lon_20hz', 'lat_20hz', 'f1', 'time_20hz']
2025-05-14 10:59:49 WARNING NaN values in CLIP 'is_bounded(-40, deg_normalize(-90, lat_20hz), 60) && is_bounded(-50, deg_normalize(-180, lon_20hz), 150)' evaluation: the result of the condition may not be the expected one. Invalid values (NaN) are rejected.
2025-05-14 10:59:50 INFO Computing done.
Insert new customized fields:
[7]:
dsx = cd_ncdf.data
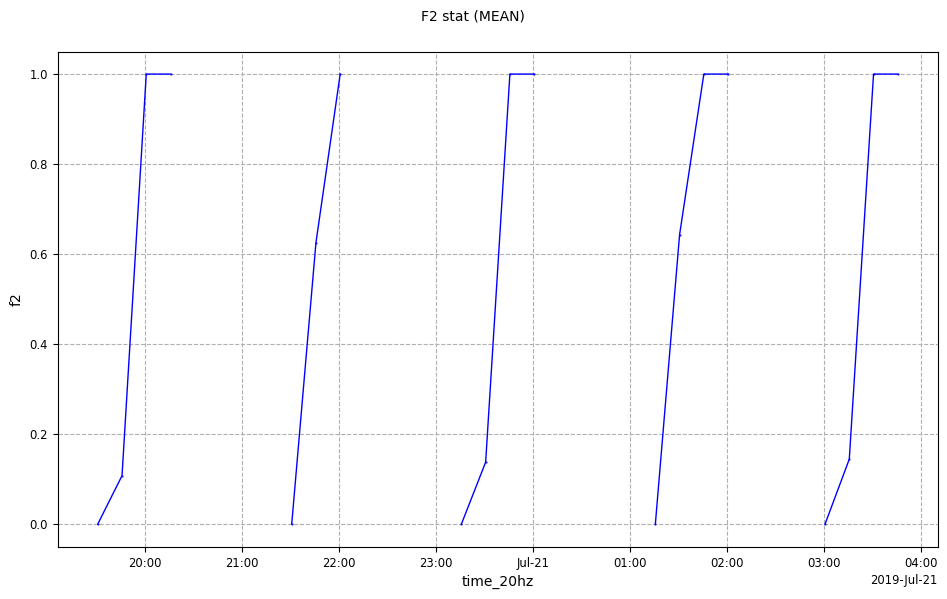
flag = xr.where(dsx["lat_20hz"] > 0, 1, 0)
dsx["f2"] = flag
cd_ncdf.data = dsx
Use these new fields:
[8]:
f3 = Field("f3", source="IIF(f2:==1, f1, DV)")
cd_ncdf.add_raw_data(name="F3", field=f3)
cd_ncdf.add_time_stat(
name="F1 stat", field=f1, freq="15min", stats=["mean", "std", "count"]
)
cd_ncdf.add_time_stat(
name="F2 stat", field=Field("f2"), freq="15min", stats=["mean", "std", "count"]
)
cd_ncdf.compute()
2025-05-14 10:59:50 INFO Reading ['f3'] (external source)
2025-05-14 10:59:50 INFO Computing diagnostics ['F1 stat', 'F2 stat']
2025-05-14 10:59:50 INFO Computing done.
Visualize everything:
[9]:
cp1 = CasysPlot(
data=cd_ncdf, data_name="F3", plot_params=PlotParams(grid=True), plot="map"
)
cp1.show()
[9]:

[10]:
cp2 = CasysPlot(
data=cd_ncdf, data_name="F2 stat", stat="mean", plot_params=PlotParams(grid=True)
)
cp2.show()
[10]: