Crossovers diagnostics - Swath
add_crossover_stat()
method of the SwathData() container.field.
the difference between the
fieldvalues of the two arcsthe difference between the
timevalues of the two arcs
field values of the two
arcs:
geographical box diagnostics using the parameters:
statsorgeobox_stats
res_lon
res_lat[optional] along time diagnostics using the parameters:
statsortemporal_stats
temporal_stats_freqtemporal_freq_kwargs: dictionary of additional parameters to pass to the pandas.date_range underlying function
Note
stats parameter is used in instance where the statistics to compute for
temporal and geobox sub-diagnostics are the same.geobox_stats and/or temporal_stats are also defined, their value will
be used in priority.diamond_reduction, diamond_relocation, cartesian_plane and
crossover_table parameters, add_crossover_stat()
allows to customize the search for crossovers and reformat the results.
pass_multi_intersect: whether to look for multiple intersections between a set of two passes or not.
cartesian_plane: flag determining the plane used for crossovers computation.
crossover_table: the table of possible combinations of crossovers between the two passes.
Add the computation of the difference between the ascending and
descending arc values in the crossovers diamonds or at crossovers
equivalent Nadir points (see diamond_reduction parameter for a
statistical reduction of the diamond data). Temporal statistics (by
cycle or day) can be added to the computation.
Values and time delta are computed at each point and requested statistics for
each geographical box. These data are accessible using the requested statistics
name or 'crossover' and 'value' keywords for the time delta and the values at
each crossover point.
Crossovers data and plots can be accessed or created using special keywords:
* delta parameter: cartographic representation of the difference
between the two arcs
* delta="field": difference of the field values
* delta="time": difference of the time values
* stat parameter: geographical box or temporal statistic representation.
* stat="...": requested statistic
* freq parameter: temporal statistic representation.
* freq="...": frequency of the requested statistic
Parameters
----------
name
Name of the diagnostic.
field
Field for which to compute the statistic.
data
External data (NadirData) to compute crossovers with.
This option is used to compute multi-missions crossovers.
max_time_difference
Maximum delta of time between the two arcs as a string with its unit.
Any string accepted by pandas.Timedelta is valid.
i.e. '10 days', '6 hours', '1 min', ...
stats
List of statistics to compute (count, max, mean, median, min, std, var,
mad) for temporal and geobox statistics.
res_lon
Minimum, maximum and box size over the longitude (Default: -180, 180, 4).
res_lat
Minimum, maximum and box size over the latitude (Default: -90, 90, 4).
box_selection
Field used as selection for computation of the ``count`` statistic.
Box in which the box_selection field does not contain any data will be set
to NaN instead of 0.
geobox_stats
Statistics included in the geobox diagnostic.
temporal_stats_freq
List of temporal statistics frequencies to compute.
temporal_stats
Statistics included in the temporal diagnostic.
temporal_freq_kwargs
Additional parameters to pass to pandas.date_range underlying function.
pass_multi_intersect
Whether to look for multiple intersections between a set of 2 passes or not.
cartesian_plane
Flag determining the plane used for crossovers computation.
If True, the crossover is calculated in the cartesian plane.
If False, the crossover is calculated in the spherical plane.
Defaults to True.
crossover_table
The table of possible combinations of crossovers between the two passes.
If this table is not defined, all crossovers between odd and even passes
will be calculated.
diamond_relocation
Flag determining data relocation to nadir crossover coordinates for
the statistics computation.
If True, data relocation is performed. Default value is False.
diamond_reduction
Statistic type used to reduce the data on the crossover diamond.
Reduction is disabled with the "none" value.
(Default value is "mean")
Raises
------
AltiDataError
If a data already exists with the provided name.
raw data diagnostics map plots for time and field differences
along time diagnostics plots
Mono-mission Swath/Swath crossovers
Diagnostic setting
swh computing a geographical box diagnostics
with default parameters and an along time diagnostics at
day and ‘one hour’ frequencies.mean and count statistics (defined with stats) are
computed for both sub diagnostics.mean and count statistics (defined with
geobox_stats) are computed for the geobox sub diagnostic, and the std and
count statistics (defined with temporal_stats) for the temporal sub diagnostic.temporal_freq_kwargs parameter allows to
pass additional parameters to the pandas.date_range underlying function, for custom
frequency values.diamond_relocation must be set to True.diamond_reduction parameter allows to reduce the data to the corresponding nadir coordinates,
by specifying the statistical reduction method.diamond_reduction default value is mean.
cartesian_plane: flag to determine if crossovers are computed in the cartesian place,
crossover_table: table of possible combinations of crosssovers between available passes.
from casys import Field
var_swh = Field(
name="swh_karin",
unit="m",
)
sd.add_crossover_stat(
name="Crossover SWH 1",
field=var_swh,
max_time_difference="5 days",
stats=["mean", "count"],
temporal_stats_freq=["day", "1h"],
diamond_relocation=False,
diamond_reduction="mean",
)
sd.add_crossover_stat(
name="Crossover SWH 2",
field=var_swh,
max_time_difference="5 days",
geobox_stats=["mean", "count"],
temporal_stats=["std", "count"],
temporal_stats_freq=["day", "1h"],
diamond_relocation=True,
diamond_reduction="mean",
)
sd.add_crossover_stat(
name="Crossover SWH 3",
field=var_swh,
max_time_difference="5 days",
stats=["mean", "count"],
temporal_stats_freq=["20min", "1h"],
temporal_freq_kwargs={"normalize": True},
diamond_relocation=False,
diamond_reduction="none",
)
sd.compute()
Diagnostic plotting
NadirData case, CasysPlot uses 3 parameters to determine
what to plot:
delta: plots time or field differences at crossovers
delta= “field”
delta= “time”
stat: plots geographical box diagnostics
stat+freq: plots along time diagnostics
Field and time differences at crossovers
Note
Time differences might contain more points than field differences if the field is not defined at some crossover points.
from casys import CasysPlot
plot = CasysPlot(data=sd, data_name="Crossover SWH 1", delta="time")
plot.show()

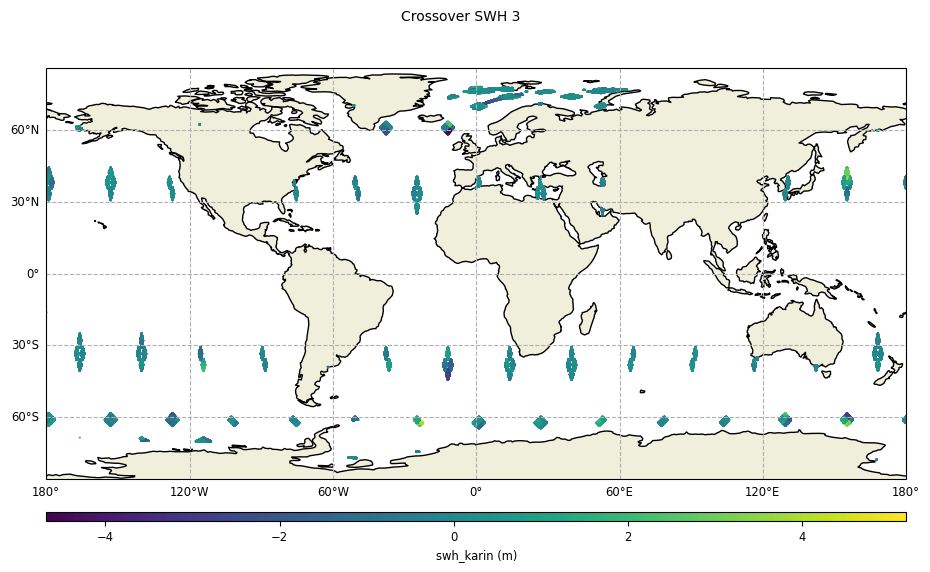
Without reduction of crossover diamonds,
diamond_reduction="none":
plot = CasysPlot(data=sd, data_name="Crossover SWH 3", delta="field")
plot.show()
from casys import PlotParams
plot = CasysPlot(
data=sd,
data_name="Crossover SWH 3",
delta="field",
plot_params=PlotParams(x_limits=(0, 30), y_limits=(-50, -25),),
)
plot.show()
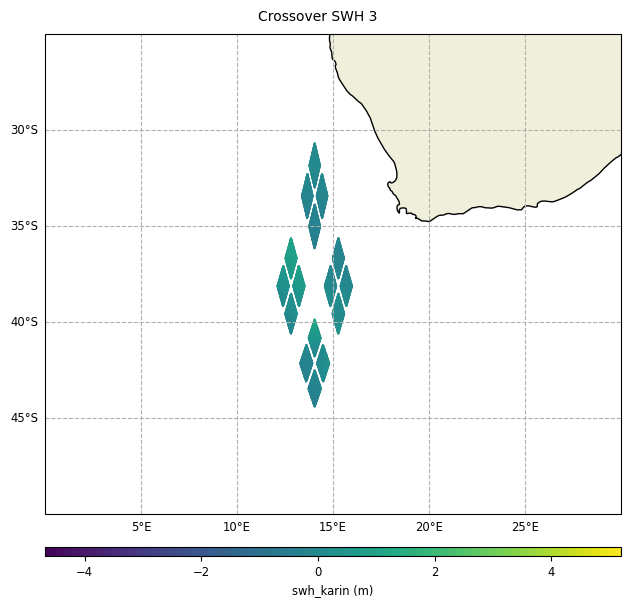
With reduction of crossover diamonds,
diamond_reduction="mean":
plot = CasysPlot(data=sd, data_name="Crossover SWH 1", delta="field")
plot.show()

plot = CasysPlot(
data=sd,
data_name="Crossover SWH 1",
delta="field",
plot_params=PlotParams(x_limits=(0, 30), y_limits=(-50, -25), marker_size=10),
)
plot.show()

Along time diagnostics
plot = CasysPlot(data=sd, data_name="Crossover SWH 1", freq="1h", stat="mean")
plot.show()

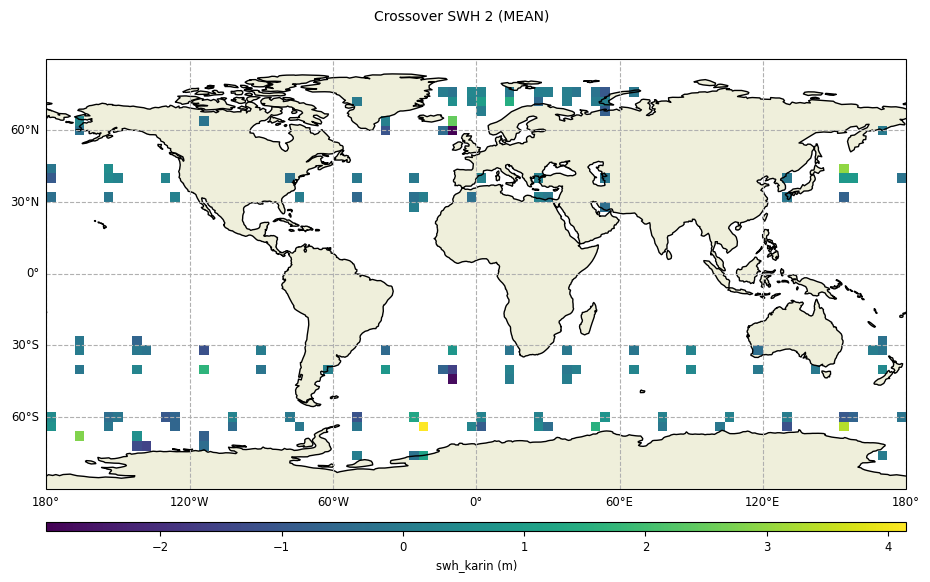
Geographical box diagnostics
diamond_relocation flag parameter value.for
diamond_relocation=False
plot = CasysPlot(data=sd, data_name="Crossover SWH 1", stat="mean")
plot.show()

for
diamond_relocation=True
plot = CasysPlot(data=sd, data_name="Crossover SWH 2", stat="mean")
plot.show()
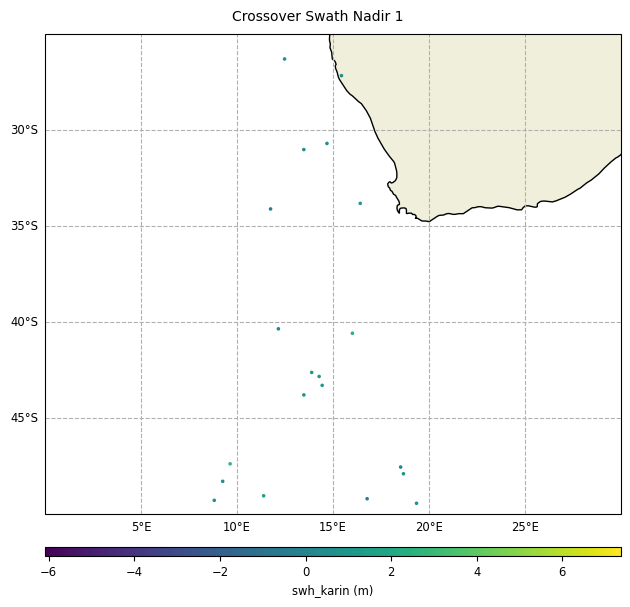
Multi-missions Swath/Nadir crossovers
data parameter of the
add_crossover_stat() method allows to
calculate crossovers with a NadirData.Diagnostic setting
Note
The following example compute crossovers between the swath type data and classic nadir data (ad):
var_swh = Field(name="swh_karin", source_aux="SWH.ALTI")
sd.add_crossover_stat(
name="Crossover Swath Nadir",
field=var_swh,
data=ad,
max_time_difference="11 days",
temporal_stats_freq=["day", "1h"],
stats=["std", "mean"],
diamond_reduction="mean",
)
sd.compute()
Diagnostic plotting
plot = CasysPlot(data=sd, data_name="Crossover Swath Nadir 1", delta="time")
plot.show()
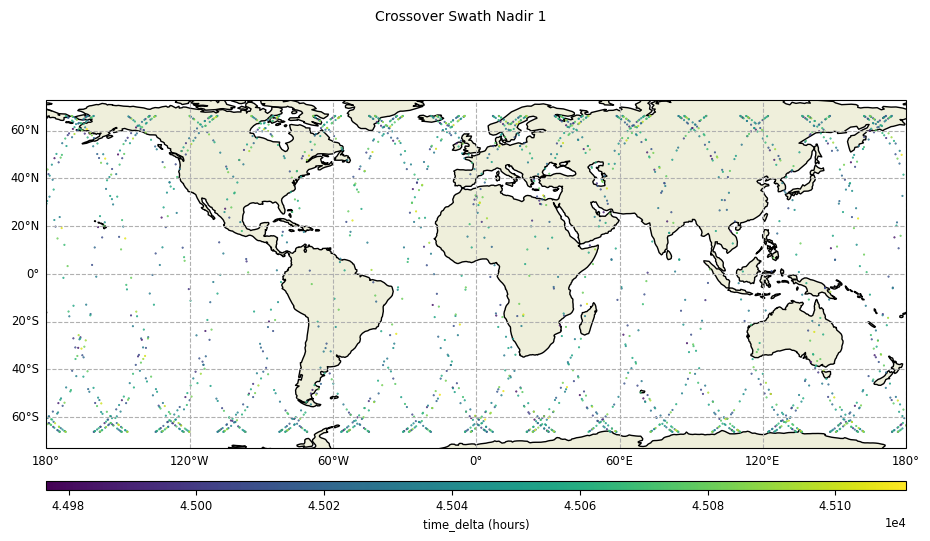
Without reduction of crossover diamonds,
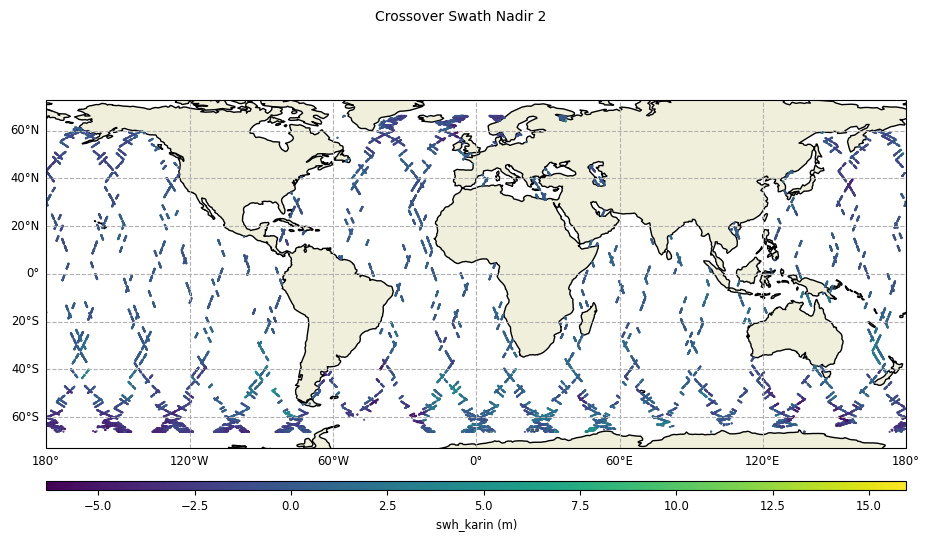
diamond_reduction="none":
plot = CasysPlot(data=sd, data_name="Crossover Swath Nadir 2", delta="field")
plot.show()
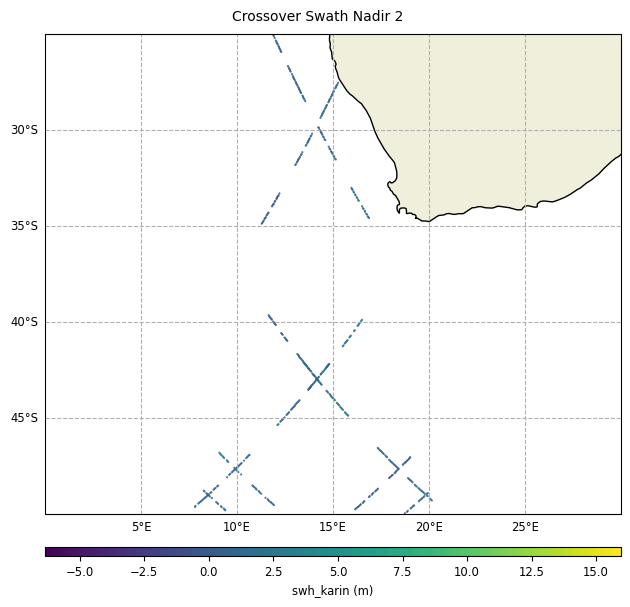
plot = CasysPlot(
data=sd,
data_name="Crossover Swath Nadir 2",
delta="field",
plot_params=PlotParams(x_limits=(0, 30), y_limits=(-50, -25)),
)
plot.show()
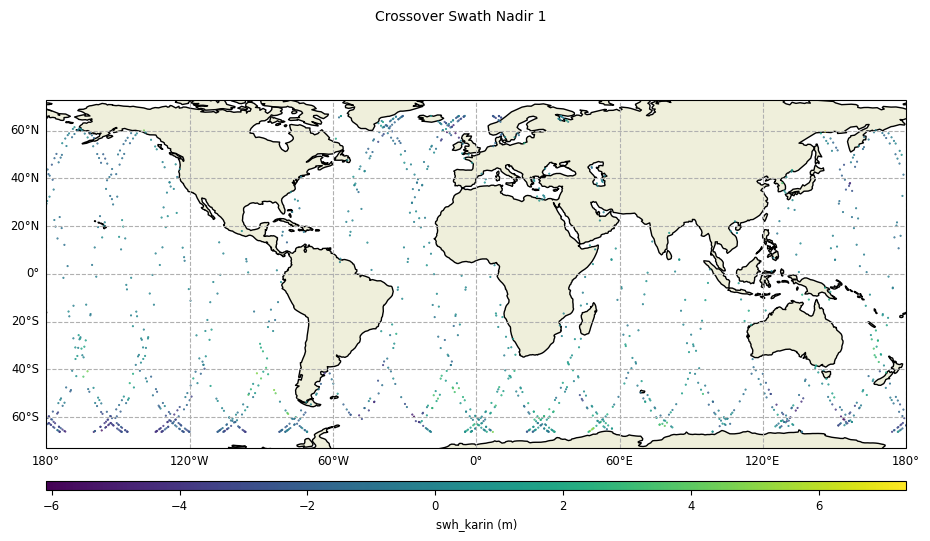
With reduction of crossover diamonds,
diamond_reduction="mean":
plot = CasysPlot(data=sd, data_name="Crossover Swath Nadir 1", delta="field")
plot.show()
plot = CasysPlot(
data=sd,
data_name="Crossover Swath Nadir 1",
delta="field",
plot_params=PlotParams(x_limits=(0, 30), y_limits=(-50, -25), marker_size=10),
)
plot.show()