Raw along track data diagnostics
Raw data are added using the
add_raw_data()
method. Fields data will be extracted alongside their time, longitude and latitude
coordinates.Warning
When using a SwathData data container, the amount of data is important, so Raw diagnostics should only be used to visualize short time periods.
Add a raw data diagnostic (used for along track plotting).
Raw data and plots can be accessed or created using special keywords:
* plot="time" (default): along time representation.
* plot="map": Cartographic representation.
* plot="3d": 3d scatter representation.
Parameters
----------
name
Name of the diagnostic.
field
Field for which to get raw data.
Raises
------
AltiDataError
If a diagnostic already exists with the provided name.
Or plotted:
as an along time plot using the option: plot=”time” when initializing the CasysPlot object (default value).
as a map plot: plot=”map” when initializing the CasysPlot object.
Diagnostic setting
In the following example, we are setting a raw diagnostic:
for an existing field
SIGMA0.ALTIin the NadirData.
sig0 = ad.fields["SIGMA0.ALTI"]
ad.add_raw_data(name="Sigma 0", field=sig0)
ad.compute()
for an existing field
swh_karinin the SwathData.
swh_karin = sd.fields["swh_karin"]
sd.add_raw_data(name="SWH Karin", field=swh_karin)
sd.compute()
Diagnostic plotting
CasysPlot requires a plot parameter to
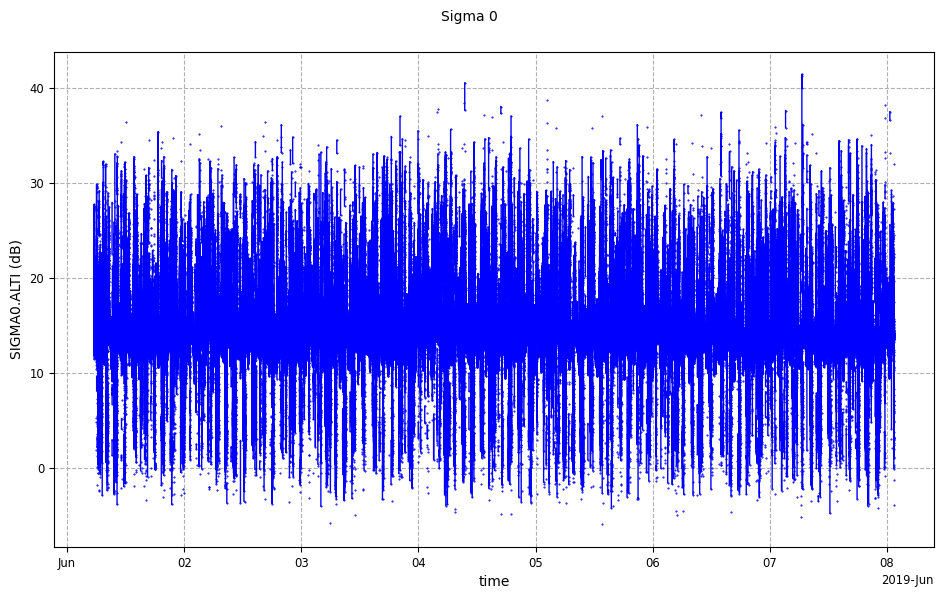
be given in order to know which plotting to use.Along time plot
Along time plots require the plot parameter to have the “time” value.
This is the default value and can omitted.
from casys import CasysPlot
plot = CasysPlot(data=ad, data_name="Sigma 0", plot="time")
plot.show()
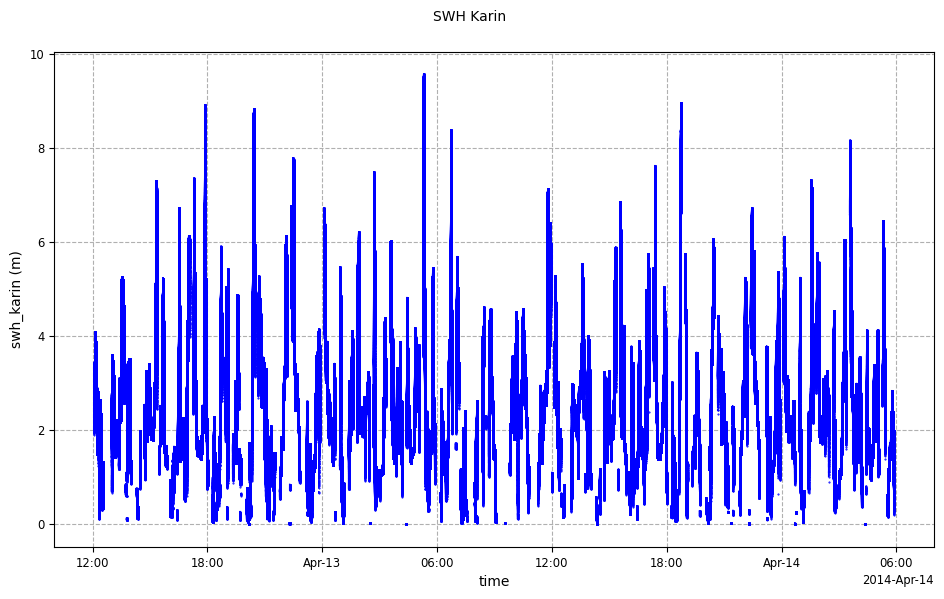
For SwathData data container, the time period to visualize might be limited in order to reduce
the quantity of points to display.
from casys import DataParams, DateHandler
time_limits = (
DateHandler("2014-04-12 15:00:00"),
DateHandler("2014-04-12 15:01:00"),
)
plot = CasysPlot(data=sd,
data_name="SWH Karin",
plot="time",
data_params=DataParams(
time_limits=time_limits,
),
)
plot.show()
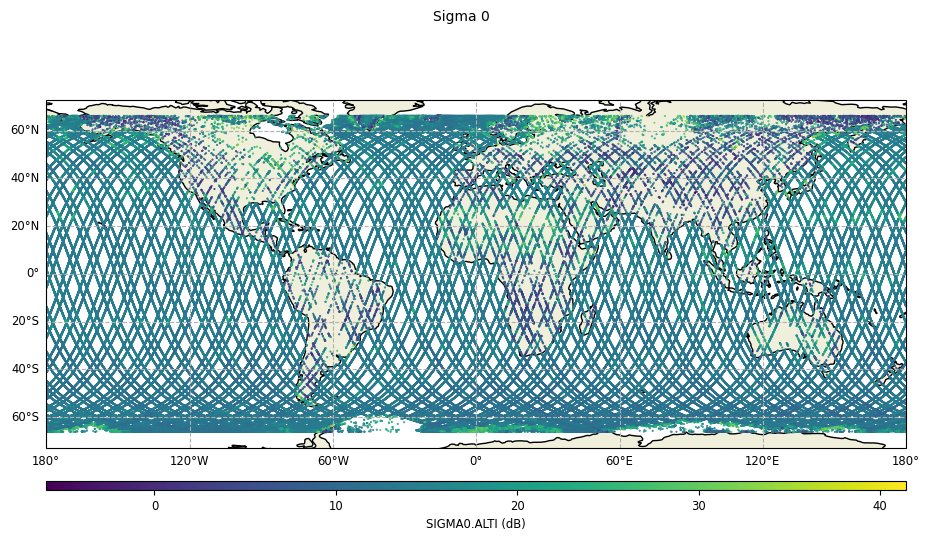
Map plot
Map plots require the plot parameter to have the “map” value.
plot = CasysPlot(data=ad, data_name="Sigma 0", plot="map")
plot.show()
You might want to use the points_min_radius parameter when visualizing swath data.
time_limits = (
DateHandler("2014-04-12 15:00:00"),
DateHandler("2014-04-12 15:00:30"),
)
plot = CasysPlot(
data=sd,
data_name="SWH Karin",
plot="map",
data_params=DataParams(
time_limits=time_limits,
points_min_radius=0.2,
),
)
plot.show()
3D plot
3D plots require the plot parameter to have the “3d” value.
NadirData containers will plot data with as a
scatter:plot = CasysPlot(data=ad, data_name="Sigma 0", plot="3D")
plot.show()

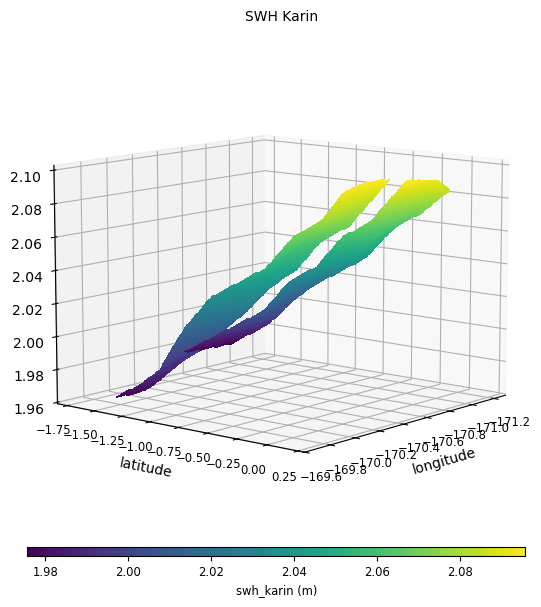
SwathData data will be plotted as
surfacesThe two swaths of the data can be split using
plot="3d:n_pix",
with n_pix the index of the splitting pixel.In the same way as 2D plot, you might want to use the
points_min_radius parameter when visualizing swath data.time_limits = (
DateHandler("2014-04-12 15:00:00"),
DateHandler("2014-04-12 15:00:30"),
)
plot = CasysPlot(
data=sd,
data_name="SWH Karin",
plot="3D:35",
data_params=DataParams(
time_limits=time_limits,
points_min_radius=0.1,
),
)
plot.show()