Spectral analysis
NadirData() or SwathData().add_spectral_analysis metonhod of the used data container.field,
by completing the following steps:Selecting valid data segments
Computing the spectral curves
segment_length: length of a segment. Needs to be provided.holes_max_length: maximum length of a hole (Default: 1% of the ``segment_length`` value).global_resampling: resampling flag (Default: False).delta_t: time gap between two measurements.noise_amplitude: noise amplitude in data (Default: half of the data standard deviation).insulation_level: minimum valid values percentage on both sides of the hole (Default: 0.75).last_segment_overlapping: percentage of overlap for second-to-last segment (Default: 0)max_time_dispersion: maximum allowed percentage of dispersion for delta_t (Default: 5).max_resampling: maximum resampled data percentage (Default: 0.25).segments_nb_delta_t: number of segments used to compute the average time gap between two measurements, during the segments extraction process (Default: 1).segments_nb_delta_x: number of segments used to compute the average distance between two measurements, during the segments extraction process (Default: 1).
segments_nb_delta_t and segments_nb_delta_x,
are used for non constant step cases.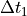 stands for the low resolution (or normal resolution) time delta, in seconds.
stands for the low resolution (or normal resolution) time delta, in seconds.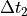 stands for the high resolution time delta, in seconds.
stands for the high resolution time delta, in seconds.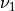 and
and 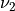 are the frequency in Hz corresponding to
are the frequency in Hz corresponding to 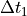 and
and 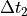 .
.

Mission |
Ground speed (km/s) |
|
|
pts nb @ |
|
|
pts nb @ |
|---|---|---|---|---|---|---|---|
Topex/Poseidon
(TP)
|
5.77 |
1.07858 |
1 |
161 |
|||
Jason1
(J1)
|
5.77 |
1.02 |
1 |
170 |
0.051 |
20 |
3398 |
Jason2 and Jason3
(J2/J3)
|
5.77 |
1.01941747572816 |
1 |
170 |
0.0509708737864078 |
20 |
3400 |
Sentinel-6
(S6A)
|
6.7 |
1 |
1 |
173 |
0.05 |
20 |
3466 |
Sentinel-3
(S3A/S3B)
|
6.7 |
1 |
1 |
149 |
0.0476190476190476 |
20 |
3134 |
SARAL/AltiKa
(AL)
|
6.73 |
1.04 |
1 |
142 |
0.026 |
40 |
5715 |
Cryosat-2
(C2)
|
6.77 |
0.9434 |
1 |
157 |
0.04717 |
20 |
3131 |
HY-2
(H2/H2B/H2C)
|
6.337 |
1.024 |
1 |
154 |
0.0512 |
20 |
3082 |
CFOSAT
(CFO)
|
7.03 |
10.857 |
0.21714 |
5 |
655 |
add_spectral_analysis() documentation.
spectral_conf: dictionary defining the parameters used to perform the spectral analysis,
res_segments: flag to indicate whether or not to keep the segments data in the results (Default:False),
res_individual_psd: flag to indicate whether or not to keep the individual power spectrum on each segments, before averaging (Default:False).
spectral_conf parameter has the following format:spectral_conf = {
"sp_analysis_name_1": {
"spectral_type": "periodogram", "param_11": param_11_value, "param_12": param_12_value, ..
},
"sp_analysis_name_2": {
"spectral_type": "periodogram", "param_21": param_21_value, "param_22": param_22_value, ...
},
...
"sp_analysis_name_n": {
"spectral_type": "welch", "param_n1": param_n1_value, "param_n2": param_n2_value, ...
},
}
spectral_analysis_name_n is the custom name of the nth spectral analysis
configuration for the psd computation.spectral_conf parameter, the spectral_type parameter is mandatory.spectral_type are periodogram and welch.spectral_conf parameter:periodogram:
{'detrend': 'linear',
'nfft': None,
'return_onesided': True,
'scaling': 'density',
'window': ('tukey', 0.05)}
welch:
{'average': 'mean',
'detrend': 'linear',
'nfft': None,
'noverlap': 0,
'nperseg': None,
'return_onesided': True,
'scaling': 'density',
'window': ('tukey', 0.05)}
window parameter, if the value is a string or tuple, it is passed to the
scipy.signal.get_window
function to generate the window values:a character chain: name of the window to use (
"hann")a tuple: name of the window to use and additional parameters (
("tukey", 0.2))
window is array_like it will be used directly as the window and its length must be equal
to segment_length value.pixels parameter, allowing to select the pixel
values to perform the spectral analysis on.
single integer/float values indicating a single pixel value:
pixlists/tuples of two integer values indicating a range of pixel values:
[pix_min, pix_max]
It is also possible to provide a single integer value for a single pixel in the spectral analysis.
Let’s see the following pixels format examples:
# Single pixel
pixels = 20
pixels = (20)
pixels = [20]
# Several pixels
pixels = [55, 60]
pixels = (55, 60)
# Single pixels range
pixels = [[55, 60]]
pixels = [(55, 60)]
pixels = ([55, 60])
pixels = ((55, 60))
# Several pixels and pixels ranges
pixels = [20, [55, 60], 49, [15, 18]]
pixels = [20, (55, 60), 49, (15, 18)]
pixels = (20, [55, 60], 49, [15, 18])
pixels = (20, (55, 60), 49, (15, 18))
single pixel: the pixel value
20,several pixels: pixel values
55and60,single pixels range: pixel values between
55and60,20and49and pixels values between55and60and between15and18.
pixels_selection and pixels_reduction, allows to choose the final results content:
the potential results reduction and the method of reduction.pixels_reduction parameter define the method of reduction, like mean or std for example.pixels_selection parameter, the different possibilities available are:none: spectral analyses are kept for each pixel valueall: reduction of all spectral analyses to a single spectral analysisrange: reduction of spectral analyses contained in ranges(pix_min, pix_max)of thepixelsparameter
pixels_selection value is range.For the previous pixels example, the results would be depending on the pixels_selection value:
pixels pixels_selection |
“none” |
“all” |
“range” |
|---|---|---|---|
20 |
1 result for 20 |
1 result for 20 |
1 result for 20 |
[55, 60] |
2 results: 55, 60 |
1 result reduced from:
55, 60
|
2 results: 55, 60 (no range here) |
[[55, 60]] |
6 results:
55, 56, 57, 58, 59, 60
|
1 result reduced from:
55, 56, 57, 58, 59, 60
|
1 result reduced from:
55, 56, 57, 58, 59, 60
|
[20, (55, 60), 49, (15, 18)] |
11 results:
20, 55, 56, 57, 58, 59, 60, 49, 15, 16, 17, 18
|
1 result reduced from:
20, 55, 56, 57, 58, 59, 60, 49, 15, 16, 17, 18
|
4 results:
- 1 for 20
- 1 reduced from: 55, 56, 57, 58, 59, 60
- 1 for 49
- reduced from: 15, 16, 17, 18
|
res_segments=True).add_spectral_analysis() documentation.Nadir Data
add_spectral_analysis() method.Add the computation of a spectral analysis diagnostic.
Spectral analysis data and plots can be accessed or created
using special keywords:
* plot="psd" (default): Power spectral density along the wave number,
* plot="segments": Cartographic representation of the selected segments.
In the plot="psd" case, a "spectral_name" parameter must be provided to
specify the required spectral analysis. If not, data of the first spectral
analysis will be returned.
The ``segments_reduction`` parameter also needs to be provided if more than
one reduction was requested or if computed using dask:
* segments_reduction="mean"
* segments_reduction="median"
Additional plotting options are available to the "psd" plot type:
* individual: setting it to ``True`` (Default: ``False``) display the set of
psd on each segments instead of the average psd,
* n_bins_psd: integer determining the number of bins along the psd values
axis for the individual=True case (Default: 100),
* second_axis: flag allowing the display of the second x-axis, for the
segment length values equivalent to the wave number.
Parameters
----------
name
Name of the diagnostic.
field
Field on which to compute the analysis.
segment_length
Length of a segment (section) in number of points.
It should be something like a few hundred points. (Example: 500 units)
holes_max_length
Maximum length of a hole. It should be something like a few points.
(Example: 5, Default: 1% of the segment_length parameter value)
global_resampling
Resampling Flag (Default: False).
True - If one section requires to be resampled => resample all sections.
False - Resample only sections requiring a resampling.
delta_t
Time gap between two measurements.
noise_amplitude
Noise amplitude in data.
Default to half of the data standard deviation.
insulation_level
Minimum valid values percentage on both sides of the hole (Default: 0.75).
Left and right sides are equal to hole length.
last_segment_overlapping
Percentage of overlap for second-to-last segment (Default: 0.5).
When the section is divided in equal segments, the last segment might be
too short, so it will take some part of data (amount depending on this
parameter) from the previous segment.
max_time_dispersion
Maximum allowed percentage of dispersion for delta_t (Default: 5).
If delta_t dispersion exceed this threshold, a warning will be displayed.
max_resampling
Maximum resampled data percentage (Default: 0.25).
A warning will be displayed if this threshold is exceeded.
The resampling of a large amount of data can have a great impact on the
final result.
segments_nb_delta_t
Number of segments used to compute the average time gap between two
measurements, during the segments extraction process (Default: 1).
segments_nb_delta_x
Number of segments used to compute the average distance between two
measurements, during the segments extraction process (Default: 1).
spectral_conf
Dictionary of the spectral parameters to use for the spectral curve types.
Each key represents a spectral analysis name is associated with a dictionary
containing the parameters. This dictionary must contain at least the
"spectral_type" key and value. (Default: dictionary containing
the default "periodogram" parameters:
{"periodogram": {"spectral_type": "periodogram",
"window": "hann", "detrend": "linear", ...}}).
segments_reduction
List of statistic types used to reduce the spectral data across segments
(Default: mean).
res_segments
Flag indicating whether to save the segments data
in the spectral analysis result (Default: False).
True - Saving segments data.
False - Not saving segments data.
res_individual_psd
Flag indicating whether to save the individual
power spectrum data on each segments
in the spectral analysis result (Default: False).
True - Saving the individual psd data.
False - Not saving the individual psd data.
raw data diagnostics map plots for segments selected for the spectral analysis
spectral power density curves plots
Diagnostic setting
sla.from casys import Field
var_sla = Field(
name="SLA",
source="ORBIT.ALTI - RANGE.ALTI - MEAN_SEA_SURFACE.MODEL.CNESCLS15",
unit="m",
)
ad.add_spectral_analysis(
name="Spectral SLA 1",
field=var_sla,
segment_length=1024,
holes_max_length=64,
spectral_conf={
"Periodogram, Tukey (alpha=0.2)": {
"spectral_type": "periodogram",
"detrend": "linear",
"window": ("tukey", 0.2),
},
"Periodogram, Blackmanharris": {
"spectral_type": "periodogram",
"window": "blackmanharris",
},
"Welch, Tukey (alpha=0.2)": {
"spectral_type": "welch",
"detrend": "linear",
"window": ("tukey", 0.2),
},
},
res_segments=True,
res_individual_psd=True,
)
ad.add_spectral_analysis(
name="Spectral SLA 2",
field=var_sla,
segment_length=2048,
holes_max_length=64,
res_segments=True,
spectral_conf={
"Periodogram, Tukey (alpha=0.2)": {
"spectral_type": "periodogram",
"detrend": "linear",
"window": ("tukey", 0.2),
},
},
)
ad.compute()
Diagnostic plotting
CasysPlot uses the plot parameter to determine
what to plot:
plot: plots segments map or spectral power density curves
plot= “segments”
plot= “psd”
plot = “psd”, an additional parameter is available: spectral_name.spectral_conf parameter provided at the diagnostic creation.Segments plotting
segment_length= 1024
from casys import CasysPlot, DataParams
plot = CasysPlot(
data=ad,
data_name="Spectral SLA 1",
plot="segments",
data_params=DataParams(points_min_radius=0.2),
)
plot.show()

segment_length= 2048
plot = CasysPlot(
data=ad,
data_name="Spectral SLA 2",
plot="segments",
data_params=DataParams(points_min_radius=0.2),
)
plot.show()

Power Spectral Density plotting
spectral_name value among the previously provided spectral_names.second_axis parameter to True (Default: False).from casys import PlotParams
plot1 = CasysPlot(
data=ad,
data_name="Spectral SLA 1",
spectral_name="Periodogram, Tukey (alpha=0.2)",
second_axis=True,
)
plot1.add_stat_bar()
plot2 = CasysPlot(
data=ad,
data_name="Spectral SLA 1",
plot="psd",
spectral_name="Periodogram, Blackmanharris",
)
plot2.add_stat_bar()
plot3 = CasysPlot(
data=ad,
data_name="Spectral SLA 1",
plot="psd",
spectral_name="Welch, Tukey (alpha=0.2)",
)
plot3.add_stat_bar()
plot1.add_plot(plot2)
plot1.add_plot(plot3)
plot1.show()

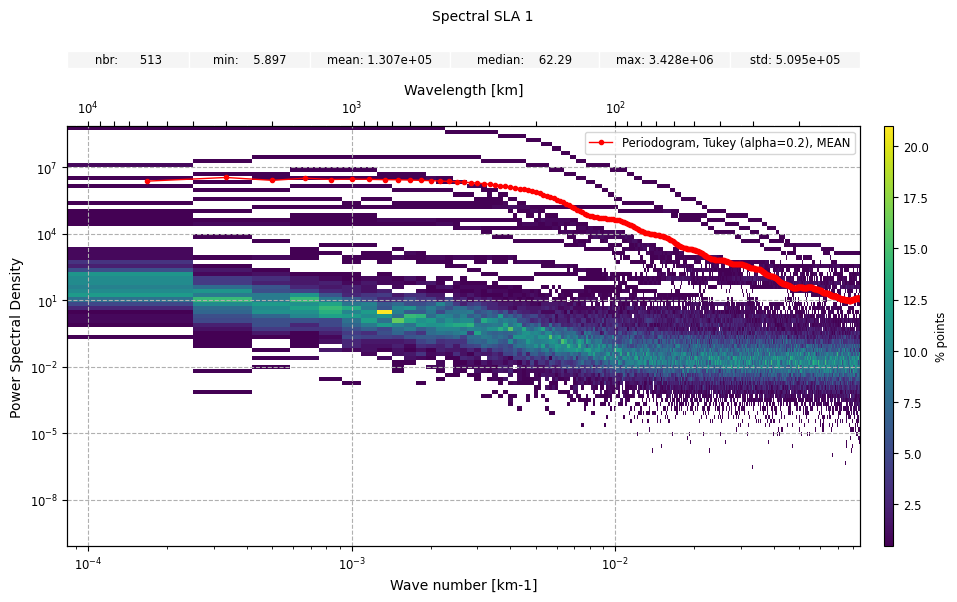
individual set to True.n_bins_psd, will determine the number
of bins along the psd values axis (Default: 100).plot1 = CasysPlot(
data=ad,
data_name="Spectral SLA 1",
plot="psd",
spectral_name="Periodogram, Tukey (alpha=0.2)",
plot_params=PlotParams(marker_style="o", marker_size=3, color="red"),
second_axis=True,
)
plot1.add_stat_bar()
plot2 = CasysPlot(
data=ad,
data_name="Spectral SLA 1",
plot="psd",
spectral_name="Periodogram, Tukey (alpha=0.2)",
individual=True,
n_bins_psd=100,
)
plot1.add_plot(plot2)
plot1.show()
Swath Data
add_spectral_analysis() method.Add the computation of a spectral analysis diagnostic.
Spectral analysis data and plots can be accessed or created
using special keywords:
* plot="psd" (default): Power spectral density along the wave number,
* plot="segments": Cartographic representation of the selected segments.
The ``segments_reduction`` parameter needs to be provided if more than one
reduction was requested or if computed using dask (the ``stat`` keyword might
be used instead):
* segments_reduction="mean"
* segments_reduction="median"
The ``pixel`` parameter needs to be provided to indicate which pixel to use:
* pixel=20
* pixel=(55, 60)
Additional plotting options are available to the "psd" plot type:
* individual: setting it to ``True`` (Default: ``False``) display the set of
psd on each segments instead of the average psd,
* n_bins_psd: integer determining the number of bins along the psd values
axis for the individual=True case (Default: 100),
* second_axis: flag allowing the display of the second x-axis, for the
segment length values equivalent to the wave number.
Parameters
----------
name
Name of the diagnostic.
field
Field on which to compute the analysis.
segment_length
Length of a segment (section) in number of points.
It should be something like a few hundred points. (Example: 500 units)
holes_max_length
Maximum length of a hole.
It should be something like a few points (Example: 5)
global_resampling
Resampling Flag (Default: False).
True - If one section requires to be resampled => resample all sections.
False - Resample only sections requiring a resampling.
delta_t
Time gap between two measurements.
noise_amplitude
Noise amplitude in data.
Default to half of the data standard deviation.
insulation_level
Minimum valid values percentage on both sides of the hole (Default: 0.75).
Left and right sides are equal to hole length.
last_segment_overlapping
Percentage of overlap for second-to-last segment (Default: 0.5).
When the section is divided in equal segments, the last segment might be
too short, so it will take some part of data (amount depending on this
parameter) from the previous segment.
max_time_dispersion
Maximum allowed percentage of dispersion for delta_t (Default: 5).
If delta_t dispersion exceed this threshold, a warning will be displayed.
max_resampling
Maximum resampled data percentage (Default: 0.25).
A warning will be displayed if this threshold is exceeded.
The resampling of a large amount of data can have a great impact on the
final result.
segments_nb_delta_t
Number of segments used to compute the average time gap between two
measurements, during the segments extraction process (Default: 1).
segments_nb_delta_x
Number of segments used to compute the average distance between two
measurements, during the segments extraction process (Default: 1).
spectral_conf
Dictionary of the spectral parameters to use for the spectral curve types.
Each key represents a spectral analysis name is associated with a dictionary
containing the parameters. This dictionary must contain at least the
"spectral_type" key and value. (Default: dictionary containing
the default "periodogram" parameters:
{"periodogram": {"spectral_type": "periodogram",
"window": "hann", "detrend": "linear", ...}}).
segments_reduction
List of statistic types used to reduce the spectral data across segments
(Default: mean).
pixels
Pixels indexes along cross track distance dimension for which to
compute the diagnostic.
Either a single integer (for a single pixel) or a list/tuple of:
- integers: pix
- list/tuple of two integers, describing a range (pix_min, pix_max)
pixels_selection
Mode of spectral data reduction for the different spectral curves
computed for the provided pixel values (Default: "all").
"all": reduce all the spectral curves for the reduction.
"range": reduce the spectral curves within a cross track distance range.
"none": no reduction performed.
pixels_reduction
Statistic type used to reduce the spectral data across pixels
(Default: mean).
res_segments
Flag indicating whether to save the segments data
in the spectral analysis result (Default: False).
True - Saving segments data.
False - Not saving segments data.
res_individual_psd
Flag indicating whether to save the individual
power spectrum data on each segments
in the spectral analysis result (Default: False).
True - Saving the individual psd data.
False - Not saving the individual psd data.
This kind of diagnostic can generate 2 kind of plots:
raw data diagnostics map plots for segments selected for the spectral analysis
spectral power density curves plots
Diagnostic setting
swh_karin.from casys import Field
var_swh = Field(
name="swh_karin",
unit="m",
)
sd.add_spectral_analysis(
name="Spectral SWH",
field=var_swh,
segment_length=1024,
holes_max_length=64,
pixels=[20, 49, [55, 60]],
spectral_conf={
"Periodogram, Tukey (alpha=0.2)": {
"spectral_type": "periodogram",
"detrend": "linear",
"window": ("tukey", 0.2),
},
},
pixels_selection="range",
pixels_reduction="mean",
res_segments=True,
res_individual_psd=True,
)
ad.compute()
Diagnostic plotting
pixel.pixels for the diagnostic setting.Segments plotting
plot = CasysPlot(
data=sd,
data_name="Spectral SWH",
plot="segments",
pixel=20,
data_params=DataParams(points_min_radius=0.2),
)
plot.show()

Power Spectral Density plotting
spectral_name value among the previously provided spectral_names.second_axis parameter to True (Default: ``False``).from casys import TextParams
plot1 = CasysPlot(
data=sd,
data_name="Spectral SWH",
plot="psd",
spectral_name="Periodogram, Tukey (alpha=0.2)",
pixel=20,
second_axis=True,
)
plot1.add_stat_bar()
plot2 = CasysPlot(
data=sd,
data_name="Spectral SWH",
plot="psd",
spectral_name="Periodogram, Tukey (alpha=0.2)",
pixel=49,
)
plot2.add_stat_bar()
plot3 = CasysPlot(
data=sd,
data_name="Spectral SWH",
plot="psd",
spectral_name="Periodogram, Tukey (alpha=0.2)",
pixel=[55, 60],
)
plot3.add_stat_bar()
plot3.add_plot(plot2)
plot1.add_plot(plot3)
plot1.show()

individual set to True.n_bins_psd, will determine the number
of bins along the psd values axis (Default: 100).from casys import TextParams
plot1 = CasysPlot(
data=sd,
data_name="Spectral SWH",
plot="psd",
spectral_name="Periodogram, Tukey (alpha=0.2)",
pixel=20,
plot_params=PlotParams(marker_style="o", marker_size=3, color="red"),
second_axis=True,
)
plot1.add_stat_bar()
plot2 = CasysPlot(
data=sd,
data_name="Spectral SWH",
plot="psd",
spectral_name="Periodogram, Tukey (alpha=0.2)",
pixel=20,
individual=True,
n_bins_psd=100,
)
plot1.add_plot(plot2)
plot1.show()
