Raw comparison diagnostics
Raw data can be compared against each other using the
add_raw_comparison() method and its
associated plot.This diagnostic can be used to compare two
x and y fields or three x, y and z fields.Add a raw data comparison.
In the case of a 3d Raw comparison (z parameter provided), data and plots
can be accessed or created using special keywords:
* plot="2d" (default): 2d scatter representation.
* plot="3d": 3d scatter representation.
Parameters
----------
name
Name of the diagnostic.
x
Field used for the x-axis.
y
Field used for the y-axis.
z
Field used for the z-axis. Optional
Raises
------
AltiDataError
If a diagnostic already exists with the provided name.
Diagnostic setting
In the following example we are setting a raw comparison diagnostic allowing to
visualize:
SLAvalues in function of theirlatitude, for the NadirData.
sla = Field(
name="SLA",
source="ORBIT.ALTI - RANGE.ALTI - MEAN_SEA_SURFACE.MODEL.CNESCLS15",
unit="m",
)
latitude = ad.fields["LATITUDE"]
ad.add_raw_comparison(name="Sla / Latitude", x=latitude, y=sla)
ad.compute()
SWHvalues in function of thecross_track_distanceandtime, for a SwathData.
ctd = sd.fields["cross_track_distance"]
time = sd.fields["time"]
sd.add_raw_comparison(
name="SWH vs time & cross track distance",
x=time,
y=ctd,
z=swh_karin,
)
sd.compute()
Diagnostic plotting
2D plot
(x, y) raw comparison diagnostics are plotted as 2D plots and do not
accept any plot parameter:
from casys import CasysPlot
plot = CasysPlot(data=ad, data_name="Sla / Latitude")
plot.show()

For (x, y, z) raw comparison diagnostics, the default plot parameter value is 2d:
from casys import DataParams, DateHandler
x_limits = (
DateHandler("2014-04-12 15:00:00"),
DateHandler("2014-04-12 15:01:00"),
)
plot = CasysPlot(
data=sd,
data_name="SWH vs time & cross track distance",
data_params=DataParams(x_limits=x_limits),
)
plot.show()

3D plot
(x, y, z) raw comparison diagnostics have a 3d plotting option available:
x_limits = (
DateHandler("2014-04-12 15:00:00"),
DateHandler("2014-04-12 15:01:00"),
)
plot = CasysPlot(
data=sd,
data_name="SWH vs time & cross track distance",
plot="3d:35",
data_params=DataParams(x_limits=x_limits),
)
plot.show()
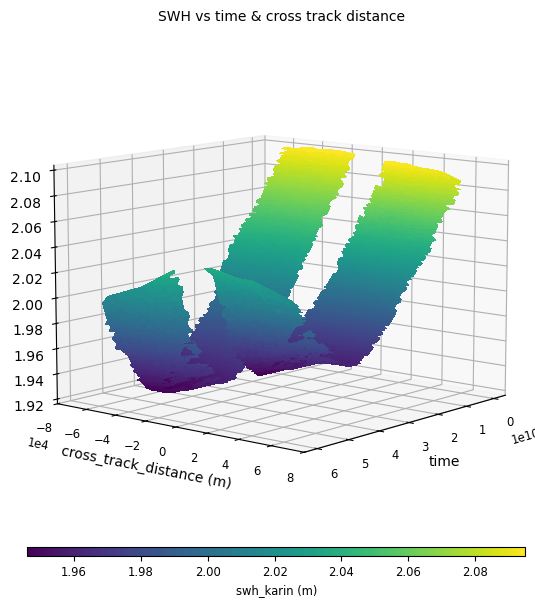
In the same way as raw diagnostics, the two swaths of the data can be split using
plot="3d:n_pix",
with n_pix the index of the splitting pixel.