Click here to download this notebook.
Crossover Swath
[2]:
import numpy as np
import swot_calval.io
from casys.readers import ScCollectionReader
from octantng.core.dask import DaskCluster
from casys import CasysPlot, DataParams, PlotParams, SwathData
SwathData.enable_loginfo()
[3]:
dask_cluster = DaskCluster(cluster_type=cluster_type, jobs=(10, 10), memory=4)
dask_client = dask_cluster.client
dask_cluster.wait_for_workers(n_workers=3, timeout=600)
Reader and container definition
Load the crossover table:
[4]:
crossover_table = swot_calval.io.load_crossover_table(
"/data/OCTANT_NG/tests/ce_swot/swot_science_2015.parquet",
latitude=80,
percentage_of_land=15,
)
Initialize the collection reader:
[5]:
collection_path = "/data/OCTANT_NG/tests/ce_swot/zcollection/"
variables = [
"cross_track_distance",
"cycle_number",
"latitude_nadir",
"latitude",
"longitude_nadir",
"longitude",
"pass_number",
"time",
"ssh_karin",
"swh_karin",
]
reader = ScCollectionReader(
data_path=collection_path,
backend_kwargs={
"cycle_numbers": 10,
"pass_numbers": np.arange(100),
"selected_variables": variables,
},
time="time",
latitude="latitude",
longitude="longitude",
latitude_nadir="latitude_nadir",
longitude_nadir="longitude_nadir",
cycle_number="cycle_number",
pass_number="pass_number",
)
[6]:
sd = SwathData(source=reader)
Definition of the crossover diagnostic
Using the add_crossover_stat method:
[7]:
sd.add_crossover_stat(
name="SSH Crossover",
field=sd.fields["ssh_karin"],
stats=["mean", "std"],
temporal_stats_freq=["1h", "DAY"],
max_time_difference="10 days",
crossover_table=crossover_table,
diamond_relocation=True,
diamond_reduction="mean",
)
[8]:
sd.add_crossover_stat(
name="SWH Crossover",
field=sd.fields["swh_karin"],
stats=["mean", "std"],
temporal_stats_freq=["1h", "DAY"],
max_time_difference="10 days",
crossover_table=crossover_table,
diamond_relocation=True,
diamond_reduction="mean",
)
[9]:
sd.add_crossover_stat(
name="SWH Crossover no relocation",
field=sd.fields["swh_karin"],
stats=["mean", "std"],
temporal_stats_freq=["1h", "DAY"],
max_time_difference="10 days",
crossover_table=crossover_table,
diamond_relocation=False,
diamond_reduction="mean",
)
Note The reduction step occurs after computing binning diagnostics.
Computation
The two crossover diagnostic on ssh_karin and swh_karin values will be computed simultaneously (same computation parameters).
[10]:
sd.compute_dask()
2025-05-14 10:58:15 INFO Computing diagnostics ['SSH Crossover']
2025-05-14 10:59:50 INFO Computing diagnostics ['SWH Crossover']
2025-05-14 11:01:05 INFO Computing diagnostics ['SWH Crossover no relocation']
2025-05-14 11:02:21 INFO Computing done.
Note Crossover diagnostics can be computed without crossover table input, at computation time cost.
Plot
Along time diagnostic
Plot the temporal diagnostic, for a specific statistic and frequency:
meanstatistic and1hfrequency forssh_karin
[11]:
plot_along_time_ssh = CasysPlot(
data=sd,
data_name="SSH Crossover",
stat="mean",
freq="1h",
data_params=DataParams(remove_nan=True),
)
plot_along_time_ssh.add_stat_bar()
plot_along_time_ssh.show()
[11]:
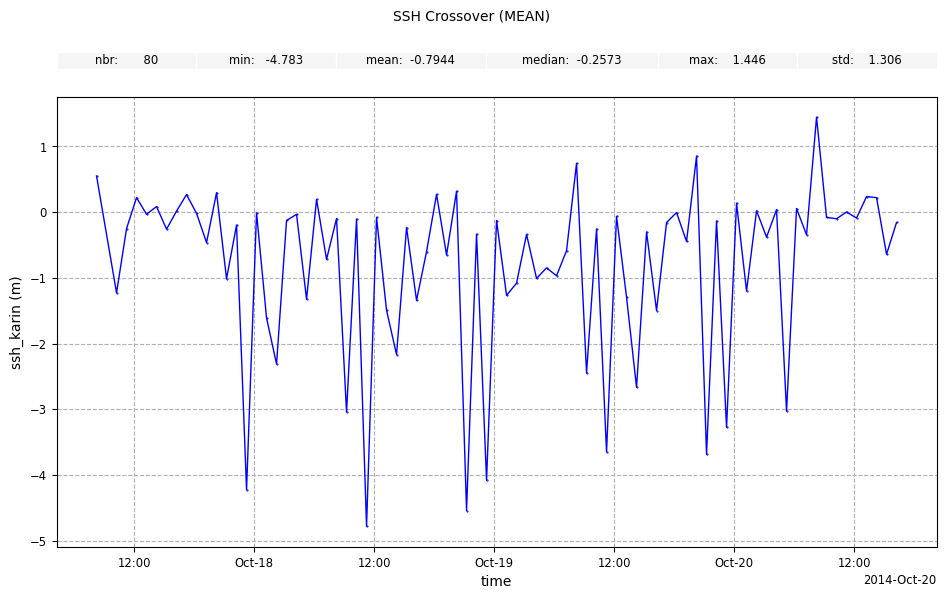
stdstatistic andDAYfrequency forswh_karin
[12]:
plot_along_time_swh = CasysPlot(
data=sd,
data_name="SWH Crossover",
stat="std",
freq="DAY",
data_params=DataParams(remove_nan=True),
)
plot_along_time_swh.add_stat_bar()
plot_along_time_swh.show()
[12]:
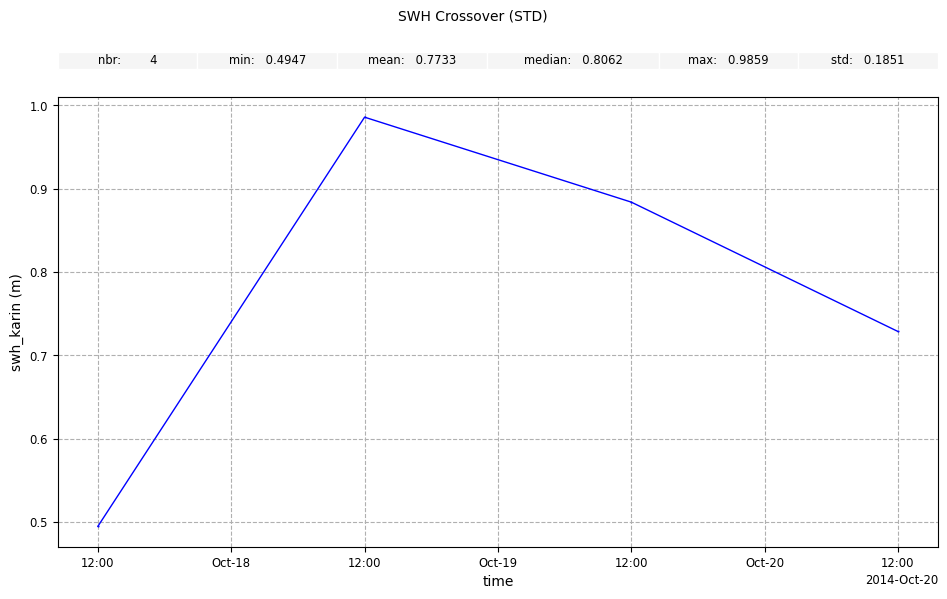
Geographical box diagnostics
Plot the geobox diagnostics, providing the diagnostic name and a specific statistic:
meanstatistic forssh_karin
[13]:
plot_geobox_mean = CasysPlot(
data=sd,
data_name="SSH Crossover",
stat="mean",
plot_params=PlotParams(color_limits=(-3, 3)),
)
plot_geobox_mean.add_stat_bar()
plot_geobox_mean.show()
[13]:
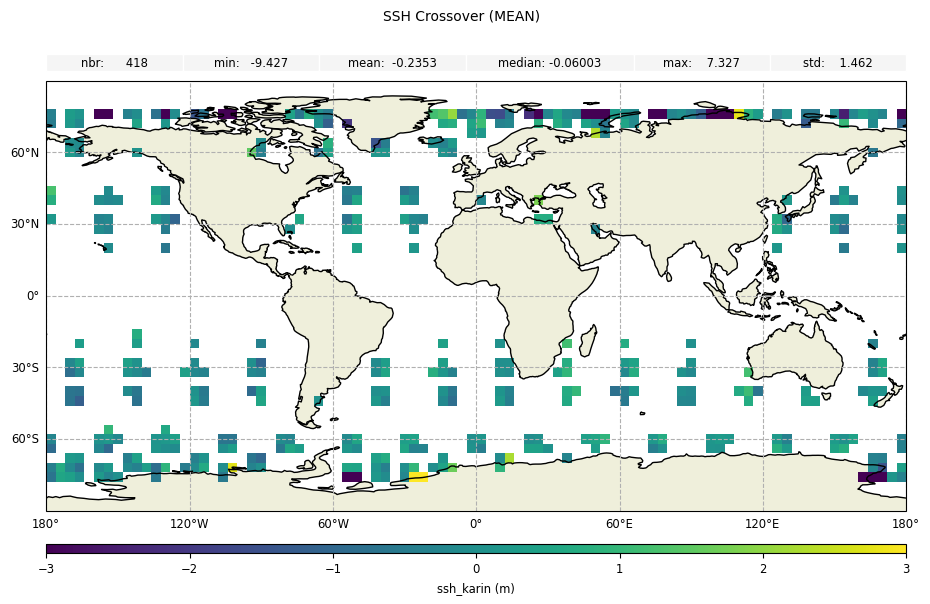
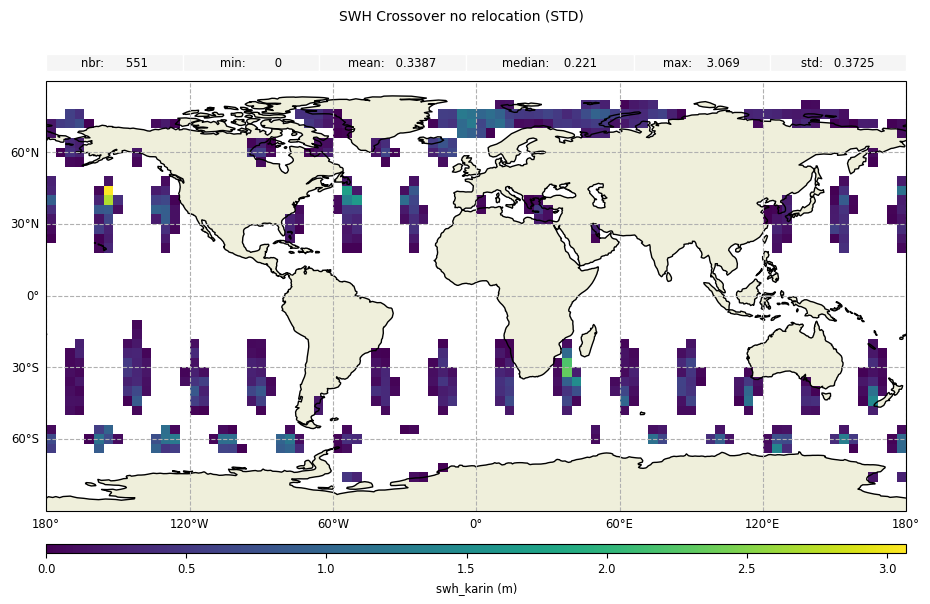
stdstatistic forswh_karin
[14]:
plot_geobox_std = CasysPlot(
data=sd,
data_name="SWH Crossover",
stat="std",
)
plot_geobox_std.add_stat_bar()
plot_geobox_std.show()
[14]:
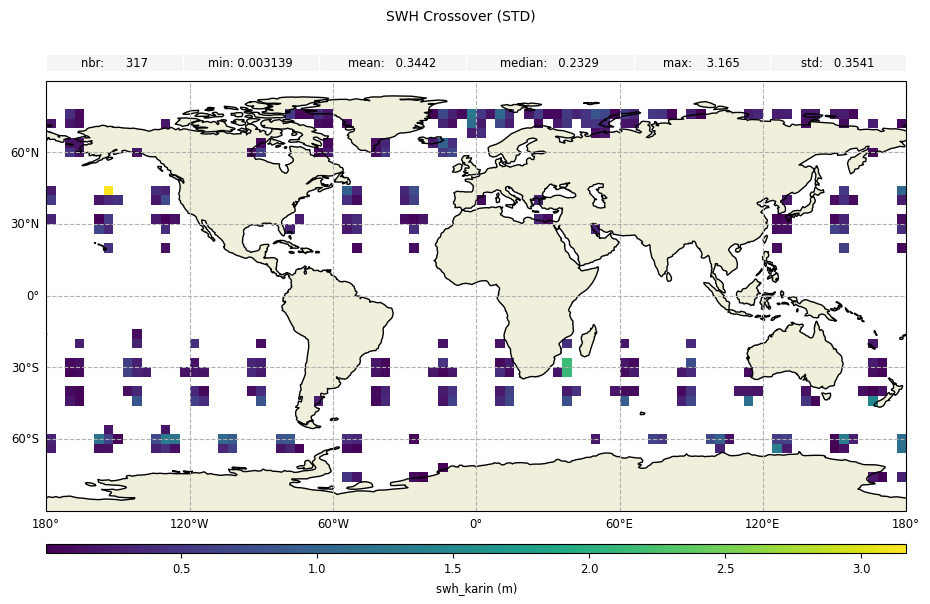
For the crossover diagnostic on swh without diamond relocation, we can notice small differences:
[15]:
plot_geobox_no_reloc = CasysPlot(
data=sd,
data_name="SWH Crossover no relocation",
stat="std",
)
plot_geobox_no_reloc.add_stat_bar()
plot_geobox_no_reloc.show()
[15]:
Field and time differences at crossovers
Plot the time or field value at crossover points (on the full diamonds or at nadir coordinates depending on the diamond_reduction parameter).
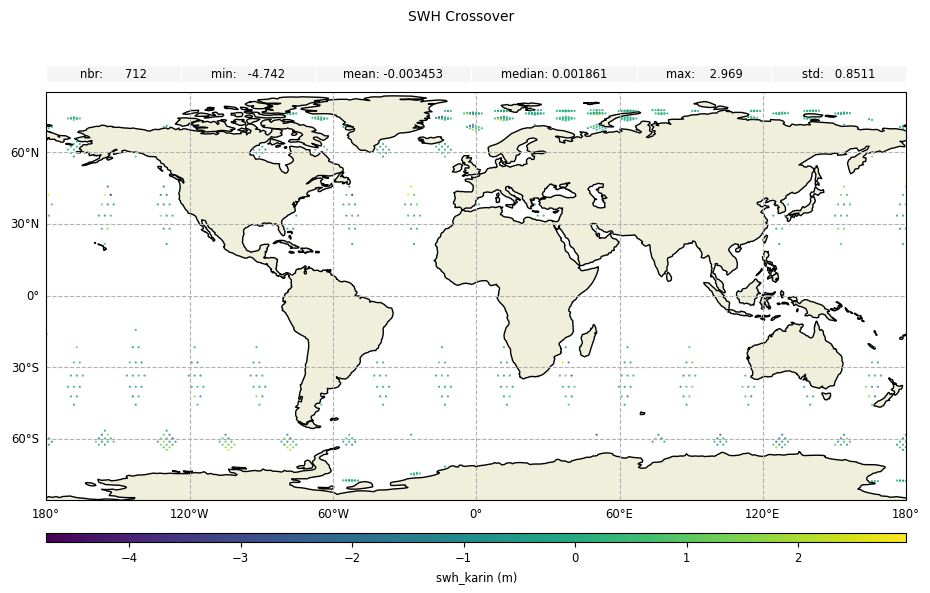
Field delta values
For field values at crossover points. Here diamond_reduction='mean', the field value is the averaged value of the field difference on the crossover diamond.
[16]:
plot_field_delta = CasysPlot(
data=sd,
data_name="SWH Crossover",
delta="field",
)
plot_field_delta.add_stat_bar()
plot_field_delta.show()
[16]:
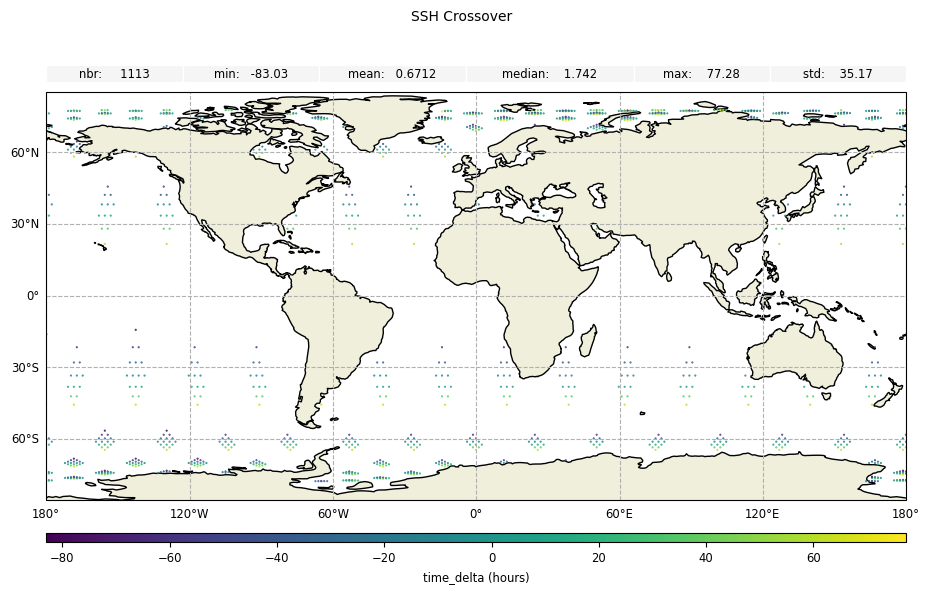
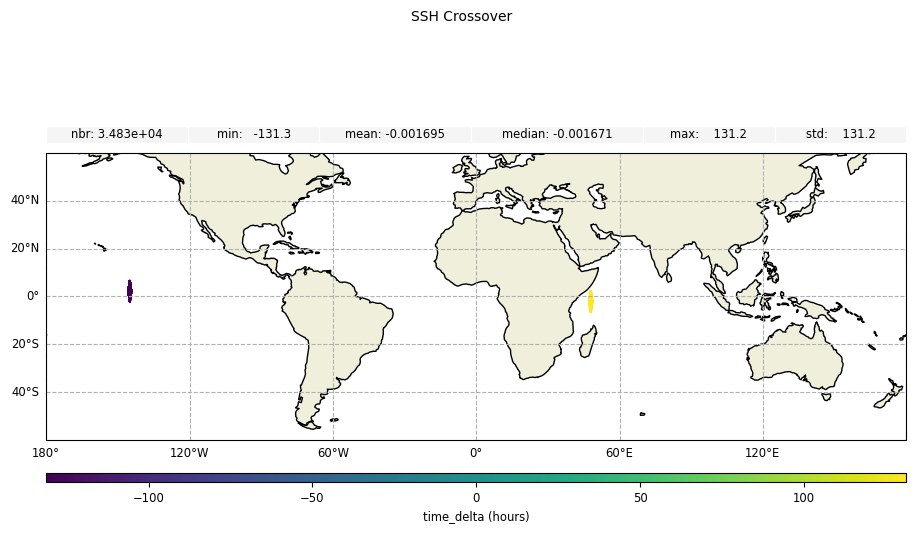
Time delta values
For time difference at crossover points:
[17]:
plot_time_delta = CasysPlot(
data=sd,
data_name="SSH Crossover",
delta="time",
)
plot_time_delta.add_stat_bar()
plot_time_delta.show()
[17]:
Example without crossovers diamond reduction
Query the collection a few passes and repeat the previous steps with diamond_reduction=False for the crossover diagnostic:
Creating SwathData object:
[18]:
reader_part = ScCollectionReader(
data_path=collection_path,
backend_kwargs={
"cycle_numbers": 10,
"pass_numbers": [1, 154, 2, 155],
"selected_variables": variables,
},
time="time",
latitude="latitude",
longitude="longitude",
latitude_nadir="latitude_nadir",
longitude_nadir="longitude_nadir",
cycle_number="cycle_number",
pass_number="pass_number",
)
[19]:
sd_part = SwathData(source=reader_part)
Adding the crossover diagnostic with
diamond_reduction=False:
[20]:
sd_part.add_crossover_stat(
name="SSH Crossover",
field=sd_part.fields["ssh_karin"],
stats="mean",
max_time_difference="10 days",
crossover_table=crossover_table,
diamond_relocation=True,
diamond_reduction="none",
)
Computing the diagnostic:
[21]:
sd_part.compute()
2025-05-14 11:02:24 INFO Computing diagnostics ['SSH Crossover']
2025-05-14 11:02:27 INFO Computing done.
Plot the field value difference for crossover diamond points:
[22]:
plot_field_delta = CasysPlot(
data=sd_part,
data_name="SSH Crossover",
delta="field",
plot_params=PlotParams(grid=True, y_limits=(-60, 60)),
)
plot_field_delta.add_stat_bar()
plot_field_delta.show()
[22]:
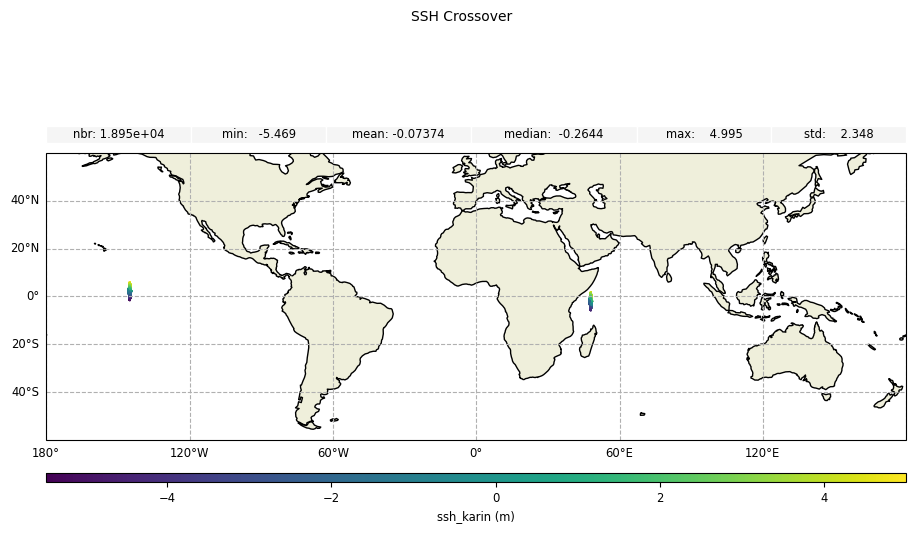
Zoom on a crossover diamond:
[23]:
plot_field_delta = CasysPlot(
data=sd_part,
data_name="SSH Crossover",
delta="field",
plot_params=PlotParams(grid=True, y_limits=(-3, 7), x_limits=(-150, -140)),
)
plot_field_delta.show()
[23]:
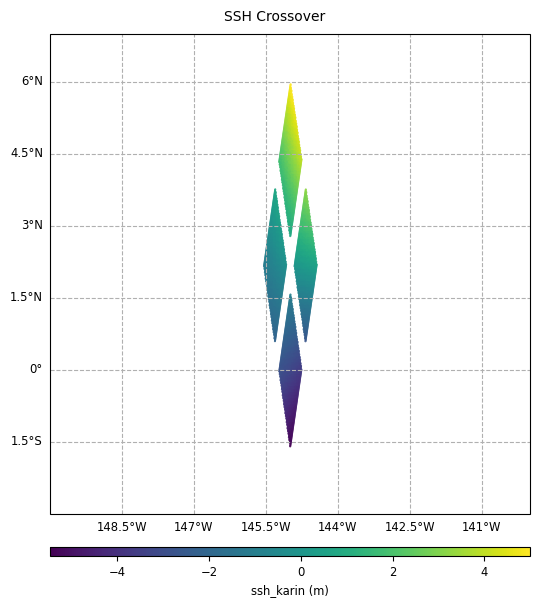
Plot the time difference value for crossover diamond points:
[24]:
plot_time_delta = CasysPlot(
data=sd_part,
data_name="SSH Crossover",
delta="time",
plot_params=PlotParams(grid=True, y_limits=(-60, 60)),
)
plot_time_delta.add_stat_bar()
plot_time_delta.show()
[24]:
Zoom on another crossover diamond:
[25]:
plot_time_delta = CasysPlot(
data=sd_part,
data_name="SSH Crossover",
delta="time",
plot_params=PlotParams(grid=True, y_limits=(-8, 4), x_limits=(35, 60)),
)
plot_time_delta.show()
[25]:

Extracting results from diagnostic
Extract the result from the SwathData object specifying the diagnostic name:
[26]:
ssh_diag = sd_part.get_diagnostic(name="SSH Crossover")
data: xarray dataset containing crossover difference, geobox binning and temporal binning results
delta: difference results at crossover diamond points (reduced to crossover nadir points if the parameter
diamond_reductionwas filled)
[27]:
ssh_diag.results.delta
[27]:
<xarray.Dataset> Size: 1MB
Dimensions: (num_lines: 34828)
Coordinates:
* time (num_lines) datetime64[ns] 279kB 2014-10-20T00:44:40.39246297...
longitude (num_lines) float64 279kB 48.35 48.37 47.94 ... -145.4 -145.4
latitude (num_lines) float64 279kB -1.203 -1.206 1.819 ... 1.318 1.32
Dimensions without coordinates: num_lines
Data variables:
ssh_karin (num_lines) float64 279kB 3.114 3.203 nan nan ... nan -1.6 nan
time_delta (num_lines) float64 279kB 131.2 131.2 131.2 ... -131.2 -131.2geo_box: geographic binning results
temporal: along time binning results
locations: structured array containing crossovers details (lon, lat, cycle_numbers, pass_numbers)
[28]:
ssh_diag.results.locations
[28]:
array([[( 47.95402409, -2.0577527 , 10, 10, 1, 154)],
[(-144.9911884 , 2.17518013, 10, 10, 155, 2)]],
dtype=[('lon', '<f8'), ('lat', '<f8'), ('cycle_number1', '<i8'), ('cycle_number2', '<i8'), ('pass_number1', '<i8'), ('pass_number2', '<i8')])
To learn more about crossovers definition, please visit this documentation page.