Click here to download this notebook.
Raw data
[2]:
from casys.readers import CLSTableReader
from casys import CasysPlot, DataParams, DateHandler, NadirData, PlotParams
NadirData.enable_loginfo()
Select your data source
[3]:
# Reader definition
table_name = "TABLE_C_J3_B_GDRD"
start = DateHandler("2019-06-01 05:30:29")
end = DateHandler("2019-06-07 05:47:33")
reader = CLSTableReader(
name=table_name,
date_start=start,
date_end=end,
time="time",
longitude="LONGITUDE",
latitude="LATITUDE",
)
# Data container definition
ad = NadirData(source=reader)
Define your diagnostic fields
[4]:
var_sig0 = ad.fields["SIGMA0.ALTI"]
Set up your diagnostic
Using the add_raw_data method:
[5]:
ad.add_raw_data("Sigma 0", var_sig0)
Compute it
[6]:
ad.compute()
2025-05-14 10:59:47 INFO Reading ['LONGITUDE', 'LATITUDE', 'SIGMA0.ALTI', 'time']
2025-05-14 10:59:49 INFO Computing done.
Visualize it
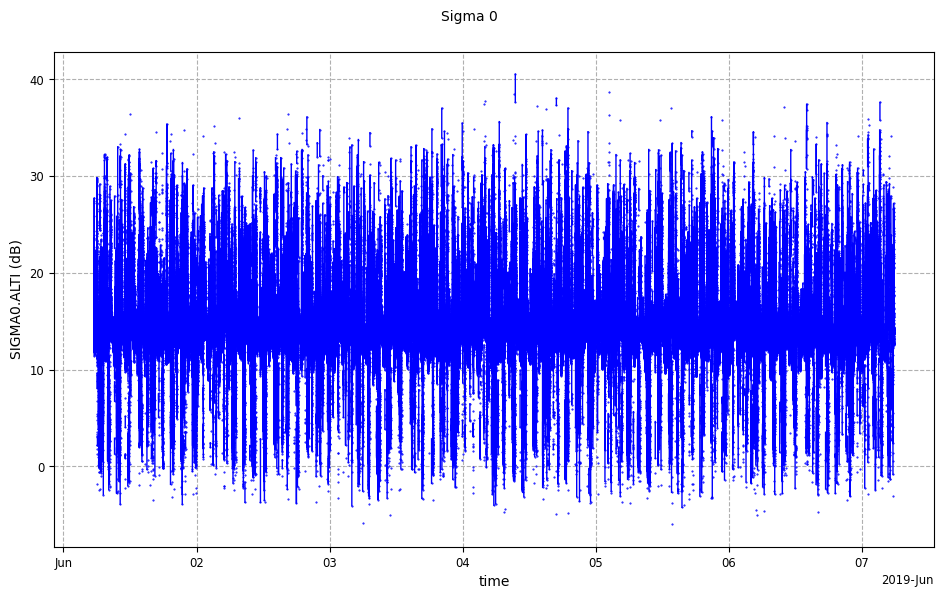
Along time plot of Sigma 0
[7]:
raw_sig0_plot = CasysPlot(ad, "Sigma 0", plot="time")
raw_sig0_plot.show()
[7]:
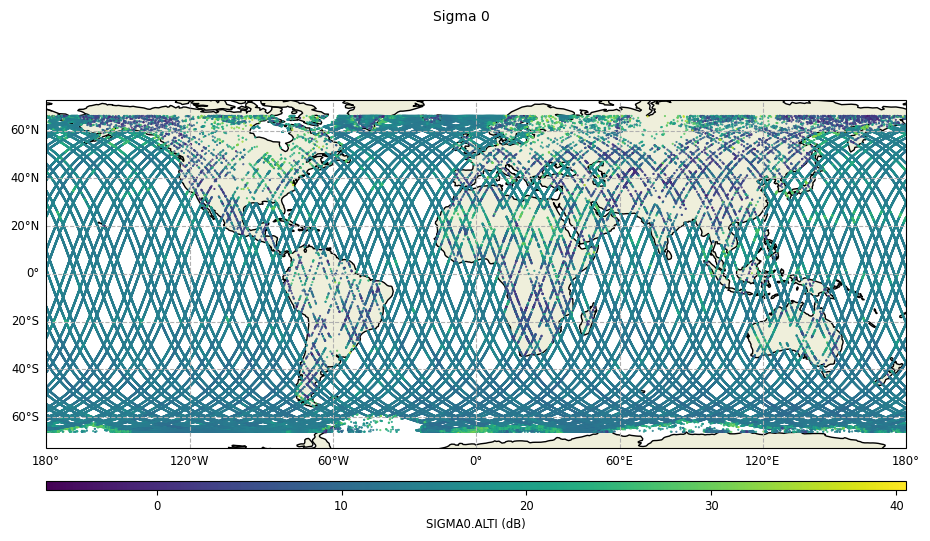
Map visualization of Sigma 0
[8]:
sig0_plot = CasysPlot(
data=ad,
data_name="Sigma 0",
plot="map",
)
sig0_plot.show()
[8]:
Plots can be customized. Here we define the color limits and y limits parameters, we display a grid and hide lands data using the grid and mask_land parameters.
[9]:
plot_par = PlotParams(
color_limits=(10, 17), y_limits=(-90, 90), grid=True, mask_land=True
)
sig0_plot = CasysPlot(data=ad, data_name="Sigma 0", plot="map", plot_params=plot_par)
sig0_plot.show()
[9]:

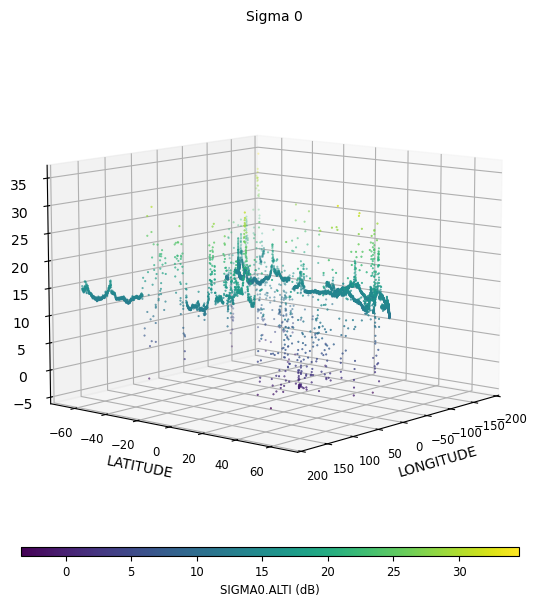
3D visualization of Sigma 0
[10]:
time_limits = ("2019-06-01 15:00:00", "2019-06-01 18:00:00")
sig0_plot = CasysPlot(
data=ad,
data_name="Sigma 0",
plot="3d",
data_params=DataParams(time_limits=time_limits),
)
sig0_plot.show()
[10]:
Additional information about Along track data can be found on this page.