Click here to download this notebook.
Crossover Swath/Nadir
[2]:
import numpy as np
from casys.readers import ScCollectionReader
from octantng.core.dask import DaskCluster
from casys import CasysPlot, Field, NadirData, PlotParams, SwathData
SwathData.enable_loginfo()
[3]:
dask_cluster = DaskCluster(cluster_type=cluster_type, jobs=(10, 10), memory=4)
dask_client = dask_cluster.client
dask_cluster.wait_for_workers(n_workers=3, timeout=600)
Swath reader and container definition
Initializing the reader:
[4]:
collection_swath_path = "/data/OCTANT_NG/tests/ce_swot/zcollection/"
variables = [
"cross_track_distance",
"cycle_number",
"latitude_nadir",
"latitude",
"longitude_nadir",
"longitude",
"pass_number",
"time",
"ssh_karin",
"swh_karin",
]
reader_swath = ScCollectionReader(
data_path=collection_swath_path,
backend_kwargs={
"cycle_numbers": 1,
"pass_numbers": np.arange(2, 16, 2),
"selected_variables": variables,
},
longitude="longitude",
latitude="latitude",
cycle_number="cycle_number",
pass_number="pass_number",
cross_track_distance="cross_track_distance",
)
Building the SwathData object:
[5]:
ad_swath = SwathData(source=reader_swath,)
Nadir reader and container definition
Initializing the reader:
[6]:
collection_nadir_path = "/data/OCTANT_NG/tests/ce_swot/zcollection_nadir"
reader_nadir = ScCollectionReader(
data_path=collection_nadir_path,
backend_kwargs={"cycle_numbers": 1, "pass_numbers": np.arange(15, 30, 2)},
)
Building the NadirData object:
[7]:
ad_nadir = NadirData(
source=reader_nadir,
time="data_01.time",
longitude="data_01.longitude",
latitude="data_01.latitude",
cycle_number="cycle_number",
pass_number="pass_number",
)
Definition of the crossover diagnostic
Using the add_crossover_stat method:
[8]:
ad_swath.add_crossover_stat(
name="SWH crossover",
field=Field(name="swh_karin", source_aux="data_01.ku.swh_ocean"),
data=ad_nadir,
stats=["mean"],
max_time_difference="5000D",
temporal_stats_freq=["1h"],
temporal_freq_kwargs={"normalize": True},
diamond_reduction="mean",
diamond_relocation=False,
)
[9]:
ad_swath.add_crossover_stat(
name="SWH crossover no reduction",
field=Field(name="swh_karin", source_aux="data_01.ku.swh_ocean"),
data=ad_nadir,
stats=["mean"],
max_time_difference="5000D",
temporal_stats_freq=["1h"],
temporal_freq_kwargs={"normalize": True},
diamond_reduction="none",
diamond_relocation=False,
)
Computation
[10]:
ad_swath.compute_dask()
2025-05-14 11:02:52 INFO Computing diagnostics ['SWH crossover']
2025-05-14 11:03:06 INFO Computing diagnostics ['SWH crossover no reduction']
2025-05-14 11:03:18 INFO Computing done.
Plot
[11]:
plot_time_delta = CasysPlot(
data=ad_swath,
data_name="SWH crossover",
delta="time",
plot_params=PlotParams(grid=True, marker_size=12),
)
plot_time_delta.show()
[11]:
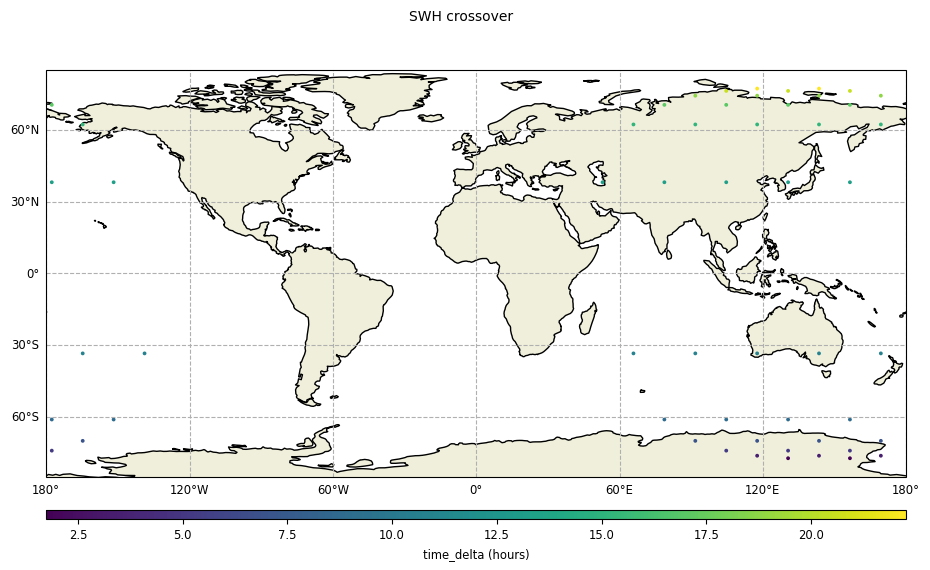
[12]:
plot_time_delta = CasysPlot(
data=ad_swath,
data_name="SWH crossover",
delta="time",
plot_params=PlotParams(
grid=True,
marker_size=12,
x_limits=(90, 150),
y_limits=(-55, -85),
),
)
plot_time_delta.show()
[12]:
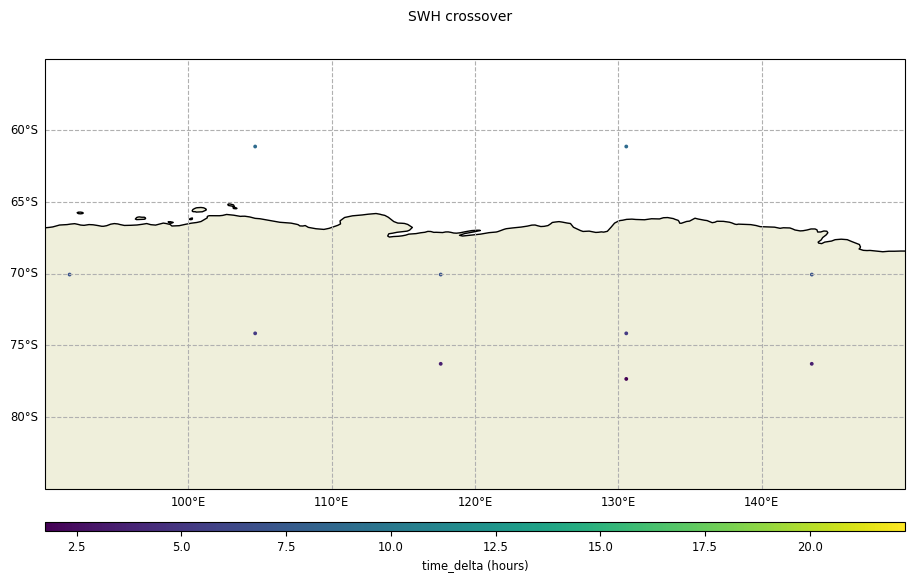
Disabling the crossover points reduction:
[13]:
plot_time_delta_no_reduction = CasysPlot(
data=ad_swath,
data_name="SWH crossover no reduction",
delta="time",
plot_params=PlotParams(grid=True),
)
plot_time_delta_no_reduction.show()
[13]:
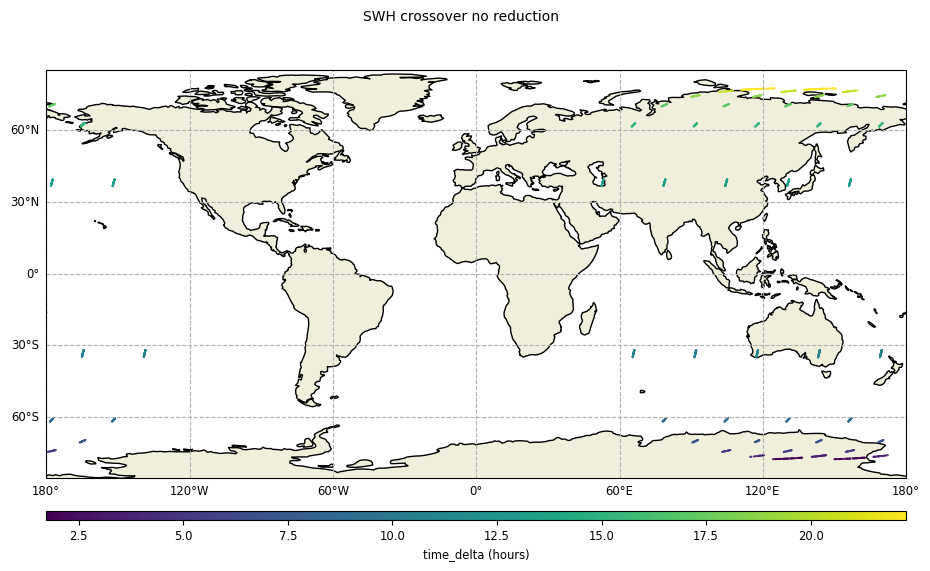
[14]:
plot_time_delta_no_reduction = CasysPlot(
data=ad_swath,
data_name="SWH crossover no reduction",
delta="time",
plot_params=PlotParams(
grid=True,
x_limits=(90, 150),
y_limits=(-55, -85),
)
)
plot_time_delta_no_reduction.show()
[14]:

Extracting results from diagnostic
Extract the result from the SwathData object specifying the diagnostic name:
[15]:
swh_diag = ad_swath.get_diagnostic("SWH crossover")
results attribute. The different type of diagnostic are:data: xarray dataset containing crossover difference, geobox binning and temporal binning results
delta: difference results at crossover diamond points (reduced to crossover nadir points if the parameter
diamond_reductionwas filled)
[16]:
swh_diag.results.delta
[16]:
<xarray.Dataset> Size: 2kB
Dimensions: (num_lines: 54)
Coordinates:
* time (num_lines) datetime64[ns] 432B 2014-04-12T18:51:34.349001504...
Dimensions without coordinates: num_lines
Data variables:
swh_karin (num_lines) float64 432B nan nan nan nan nan ... nan nan nan nan
time_delta (num_lines) float64 432B 10.82 13.24 8.827 ... 10.82 15.2 13.24
longitude (num_lines) float64 432B -138.8 -151.8 -151.8 ... 65.83 52.89
latitude (num_lines) float64 432B -33.45 38.19 -61.13 ... 62.38 38.19geo_box: geographic binning results
temporal: along time binning results
locations: structured array containing crossovers details (lon, lat, cycle_numbers, pass_numbers)
[17]:
swh_diag.results.locations
[17]:
array([[(-138.82668238, -33.4504997 , 1, 1, 2, 15)],
[(-151.77221499, 38.19113369, 1, 1, 2, 17)],
[(-164.71683509, 62.3799158 , 1, 1, 2, 19)],
[(-177.66251434, 70.58642377, 1, 1, 2, 21)],
[( 169.39406606, 74.42799932, 1, 1, 2, 23)],
[( 156.44739358, 76.42459499, 1, 1, 2, 25)],
[( 143.51175418, 77.40019387, 1, 1, 2, 27)],
[(-151.77241034, -61.12549866, 1, 1, 4, 15)],
[(-164.71694049, -33.45132379, 1, 1, 4, 17)],
[(-177.66245232, 38.19050616, 1, 1, 4, 19)],
[( 169.39220229, 62.37998941, 1, 1, 4, 21)],
[( 156.44837188, 70.58638281, 1, 1, 4, 23)],
[( 143.50376096, 74.42766592, 1, 1, 4, 25)],
[( 130.55731999, 76.42432426, 1, 1, 4, 27)],
[( 117.61335059, 77.4002118 , 1, 1, 4, 29)],
[(-164.71864158, -70.05887079, 1, 1, 6, 15)],
[(-177.66275378, -61.12619074, 1, 1, 6, 17)],
[( 169.39230358, -33.45105829, 1, 1, 6, 19)],
[( 156.44728072, 38.19071826, 1, 1, 6, 21)],
[( 143.50104311, 62.38043553, 1, 1, 6, 23)],
[( 130.55761402, 70.58624066, 1, 1, 6, 25)],
[( 117.61181823, 74.42740078, 1, 1, 6, 27)],
[( 104.66621161, 76.42426881, 1, 1, 6, 29)],
[(-177.66159746, -74.16128937, 1, 1, 8, 15)],
[( 169.39276264, -70.05872235, 1, 1, 8, 17)],
[( 156.44776748, -61.12603876, 1, 1, 8, 19)],
[( 143.50174549, -33.45093412, 1, 1, 8, 21)],
[( 130.55700962, 38.19190684, 1, 1, 8, 23)],
[( 117.6114485 , 62.38007377, 1, 1, 8, 25)],
[( 104.66588883, 70.58616269, 1, 1, 8, 27)],
[( 91.71986346, 74.42755867, 1, 1, 8, 29)],
[( 169.39139982, -76.28556809, 1, 1, 10, 15)],
[( 156.44635739, -74.16095635, 1, 1, 10, 17)],
[( 143.50302198, -70.05857956, 1, 1, 10, 19)],
[( 130.55699514, -61.12523224, 1, 1, 10, 21)],
[( 117.61153378, -33.45035182, 1, 1, 10, 23)],
[( 104.66616321, 38.19204654, 1, 1, 10, 25)],
[( 91.7219441 , 62.38032642, 1, 1, 10, 27)],
[( 78.77440312, 70.58649924, 1, 1, 10, 29)],
[( 156.44900724, -77.34060171, 1, 1, 12, 15)],
[( 143.49788588, -76.28572822, 1, 1, 12, 17)],
[( 130.55642173, -74.16048231, 1, 1, 12, 19)],
[( 117.61164932, -70.05824203, 1, 1, 12, 21)],
[( 104.66620482, -61.12512845, 1, 1, 12, 23)],
[( 91.72132102, -33.45070088, 1, 1, 12, 25)],
[( 78.77580374, 38.19093219, 1, 1, 12, 27)],
[( 65.83112887, 62.38005603, 1, 1, 12, 29)],
[( 130.55736276, -77.34022078, 1, 1, 14, 17)],
[( 117.61104008, -76.28550464, 1, 1, 14, 19)],
[( 104.66646403, -74.16042306, 1, 1, 14, 21)],
[( 91.72029973, -70.05844461, 1, 1, 14, 23)],
[( 78.77543518, -61.12577562, 1, 1, 14, 25)],
[( 65.83090233, -33.45114579, 1, 1, 14, 27)],
[( 52.88553646, 38.19046634, 1, 1, 14, 29)]],
dtype=[('lon', '<f8'), ('lat', '<f8'), ('cycle_number1', '<i8'), ('cycle_number2', '<i8'), ('pass_number1', '<i8'), ('pass_number2', '<i8')])
To learn more about crossovers definition, please visit this documentation page.