Click here to download this notebook.
Periodogram
[2]:
import numpy as np
from casys import CasysPlot, DateHandler, Field, NadirData
from casys.readers import CLSTableReader
NadirData.enable_loginfo()
Dataset definition
[3]:
# Reader definition
table_name = "TABLE_C_J3_B_GDRD"
orf_name = "C_J3"
cycle_number = 127
start = DateHandler.from_orf(orf_name, cycle_number, 1, pos="first")
end = DateHandler.from_orf(orf_name, cycle_number, 254, pos="last")
reader = CLSTableReader(
name=table_name,
date_start=start,
date_end=end,
orf=orf_name,
time="time",
longitude="LONGITUDE",
latitude="LATITUDE",
)
# Data container definition
ad = NadirData(source=reader)
var_swh = Field(name="swh", source="IIF(FLAG_VAL.ALTI==0, SWH.ALTI,DV)", unit="m")
var_sla = Field(
name="sla",
source="ORBIT.ALTI - RANGE.ALTI - MEAN_SEA_SURFACE.MODEL.CNESCLS15",
unit="m",
)
Definition of the statistics
The add_periodogram method allows the computation of periodograms from the result of a temporal diagnostic or sub-diagnostic (from a crossover or missing points diagnostic).
[4]:
first_period = np.timedelta64(1, "D")
last_period = np.timedelta64(20, "D")
nbr_periods = 64
From a temporal diagnostic
Only the stats is needed to select the correct temporal diagnostic results.
[5]:
ad.add_time_stat(name="SWH by hour", freq="1h", field=var_swh, stats=["mean", "std"])
ad.add_periodogram(
name="Periodogram SWH by hour",
base_diag="SWH by hour",
stats=["mean", "std"],
nbr_periods=nbr_periods,
first_period=first_period,
last_period=last_period,
)
From a crossover temporal sub-diagnostic
diag_kwargs, with a dictionary.diag_kwargs is the freq value, as several frequencies can be defined for the crossover temporal sub-diagnostic computation (here “cycle” and “pass”, see temporal_stats_freq).[6]:
ad.add_crossover_stat(
name="Crossover SLA",
field=var_sla,
max_time_difference="10 days",
stats=["mean", "count"],
temporal_stats_freq=["cycle", "pass"],
)
ad.add_periodogram(
name="Periodogram Crossover SLA by pass",
base_diag="Crossover SLA",
stats=["mean", "count"],
nbr_periods=nbr_periods,
first_period=first_period,
last_period=last_period,
diag_kwargs={"freq": "pass"},
)
From a missing points temporal sub-diagnostic
For a missing points temporal sub-diagnostic, the kwargs expected in diag_kwargs are:
the
freqvalue, as several frequencies can be defined for the crossover temporal sub-diagnostic computation (here “1h” and “20min”, seetemporal_stats_freq),the
dtypevalue, “missing”, “available” or “all”,the
groupvalue, “GLOBAL” or any of the other groups defined with thegroup_gridparameter (“OCEAN” and “LAND” in the default casegroup_grid=True)
[7]:
ad.add_missing_points_stat(
name="Missing Points",
reference_track="J3",
temporal_stats_freq=["1h", "20min"],
group_grid=True,
)
ad.add_periodogram(
name="Periodogram Missing Points by hour",
base_diag="Missing Points",
stats=["count"],
nbr_periods=nbr_periods,
first_period=first_period,
last_period=last_period,
diag_kwargs={"freq": "1h", "dtype": "missing", "group": "LAND"},
)
2025-05-14 10:56:13 WARNING Computed missing points will be using an ORF build with real measurements. Results might be incomplete.
Compute
[8]:
ad.compute()
2025-05-14 10:56:13 INFO Reading ['time', 'swh', 'LONGITUDE', 'LATITUDE', 'sla']
2025-05-14 10:56:17 INFO Computing diagnostics ['SWH by hour']
2025-05-14 10:56:17 INFO Computing diagnostics ['Crossover SLA']
2025-05-14 10:56:19 INFO Computing diagnostics ['Missing Points']
2025-05-14 10:56:19 INFO Searching missing points.
2025-05-14 10:56:27 INFO Computing Periodogram SWH by hour
2025-05-14 10:56:27 INFO Computing Periodogram Crossover SLA by pass
2025-05-14 10:56:27 INFO Computing Periodogram Missing Points by hour
2025-05-14 10:56:27 INFO Computing done.
[9]:
ad
[9]:
| AltiData | |||||||
|---|---|---|---|---|---|---|---|
|
Source Source type ORF Time name Longitude Latitude |
TABLE_C_J3_B_GDRD CLSTableReader C_J3 time LONGITUDE LATITUDE |
Period start Period end Selection clip Selection shape |
2019-07-20 19:23:06.823509 Cycle: 127 - Pass: 1 2019-07-30 17:21:37.233369 Cycle: 127 - Pass: 254 None False |
||||
| Time statistics | Name | Field | Frequency | Statistics | State | ||
| SWH by hour | swh | 1h | [MEAN, STD] | Computed | |||
| Periodogram analysis | Name | Base Diagnostic | Description | Statistics | State | ||
| Periodogram Crossover SLA by pass |
Crossover SLA {'freq': 'pass'} |
Number of periods: 64 First period: 1 days Last period: 20 days |
[MEAN, COUNT] | Computed | |||
| Periodogram Missing Points by hour |
Missing Points {'freq': '1h', 'dtype': MISSING, 'group': 'LAND', 'plot': 'temporal'} |
Number of periods: 64 First period: 1 days Last period: 20 days |
[COUNT] | Computed | |||
| Periodogram SWH by hour |
SWH by hour |
Number of periods: 64 First period: 1 days Last period: 20 days |
[MEAN, STD] | Computed | |||
| Crossovers | Name | Description | Fields | Resolution | Frequencies | Statistics | State |
| Crossover SLA |
Max time difference: 240.0 hours Interpolation mode: linear Spline half window size: 3 |
sla |
Longitude: min: -180, max: 180, step: 4 Latitude: min: -90, max: 90, step: 4 |
['CYCLE', 'PASS'] |
Temporal: [MEAN, COUNT] Geobox: [MEAN, COUNT] | Computed | |
| Missing Points | Name | Description | Resolution | Frequencies | State | ||
| Missing Points |
Groups: ['GLOBAL', 'LAND', 'OCEAN'] Method: REAL Distance threshold: 4.0 Reference time gap: 1.08 sec Time threshold: 1.9 |
Longitude: min: -180, max: 180, step: 4 Latitude: min: -90, max: 90, step: 4 |
['1h', '20min'] | Computed | |||
Plot
This diagnostic is plotted as curve, along the periods values computed. We need to provide a stat parameter value to the CasysPlot.
From a temporal diagnostic
[10]:
periodogram_temporal_plot = CasysPlot(
data=ad, data_name="Periodogram SWH by hour", stat="std"
)
periodogram_temporal_plot.show()
[10]:

From a crossover temporal sub-diagnostic
[11]:
periodogram_crossover_plot = CasysPlot(
data=ad, data_name="Periodogram Crossover SLA by pass", stat="mean"
)
periodogram_crossover_plot.show()
[11]:
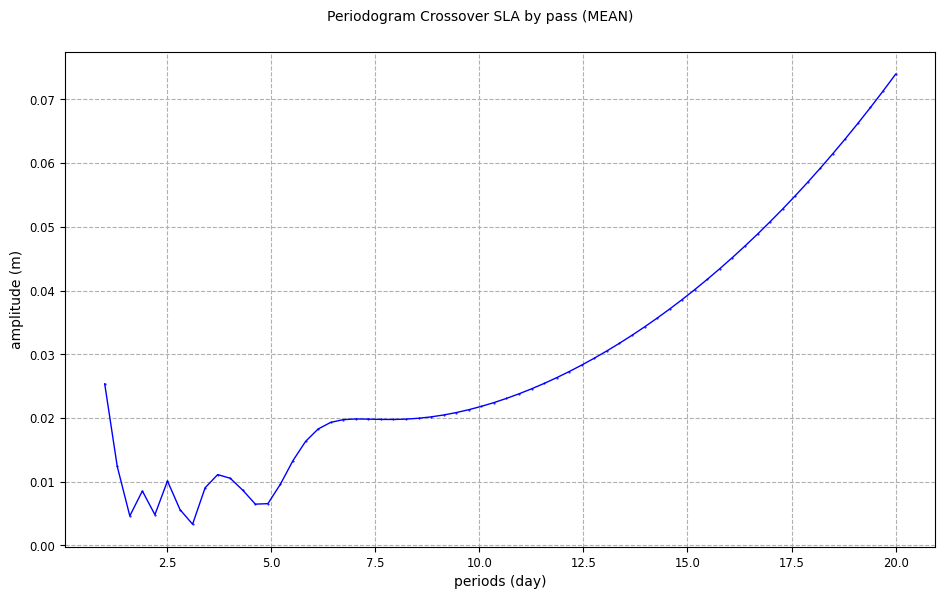
From a missing points temporal sub-diagnostic
[12]:
periodogram_missingpoints_plot = CasysPlot(
data=ad, data_name="Periodogram Missing Points by hour", stat="count"
)
periodogram_missingpoints_plot.show()
[12]:
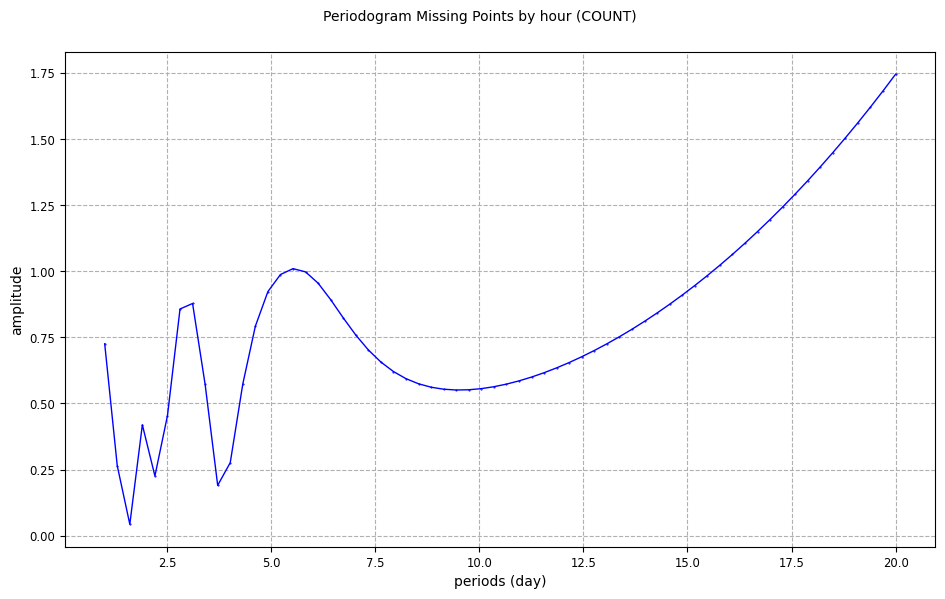
To learn more about periodogram diagnostic definition, please visit this documentation page.